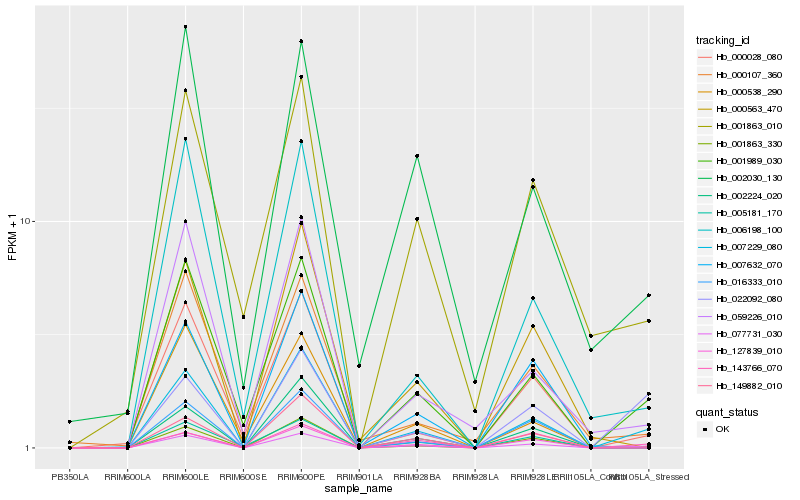
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_077731_030 |
0.0 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 2 |
Hb_007229_080 |
0.0503266977 |
- |
- |
Nonspecific lipid-transfer protein A, putative [Ricinus communis] |
| 3 |
Hb_001989_030 |
0.108048545 |
- |
- |
PREDICTED: copper transport protein ATX1 [Jatropha curcas] |
| 4 |
Hb_000028_080 |
0.1574437571 |
- |
- |
PREDICTED: uncharacterized protein At3g17950 [Jatropha curcas] |
| 5 |
Hb_002030_130 |
0.1737283555 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 6 |
Hb_006198_100 |
0.1741847424 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 7 |
Hb_059226_010 |
0.1769960236 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 8 |
Hb_127839_010 |
0.1782175786 |
- |
- |
hypothetical protein PRUPE_ppa000956mg [Prunus persica] |
| 9 |
Hb_143766_070 |
0.1796942657 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 10 |
Hb_001863_010 |
0.1813696394 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 11 |
Hb_001863_330 |
0.1837676798 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 12 |
Hb_022092_080 |
0.1867268112 |
- |
- |
- |
| 13 |
Hb_000563_470 |
0.1869574611 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 14 |
Hb_000538_290 |
0.1879939782 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 15 |
Hb_016333_010 |
0.1880839434 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 16 |
Hb_149882_010 |
0.1889788145 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Populus euphratica] |
| 17 |
Hb_002224_020 |
0.1899228583 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 18 |
Hb_005181_170 |
0.190120062 |
- |
- |
PREDICTED: aldehyde dehydrogenase 22A1 [Jatropha curcas] |
| 19 |
Hb_000107_360 |
0.1904181841 |
- |
- |
PREDICTED: xylem cysteine proteinase 1 [Jatropha curcas] |
| 20 |
Hb_007632_070 |
0.190521631 |
- |
- |
kinase, putative [Ricinus communis] |