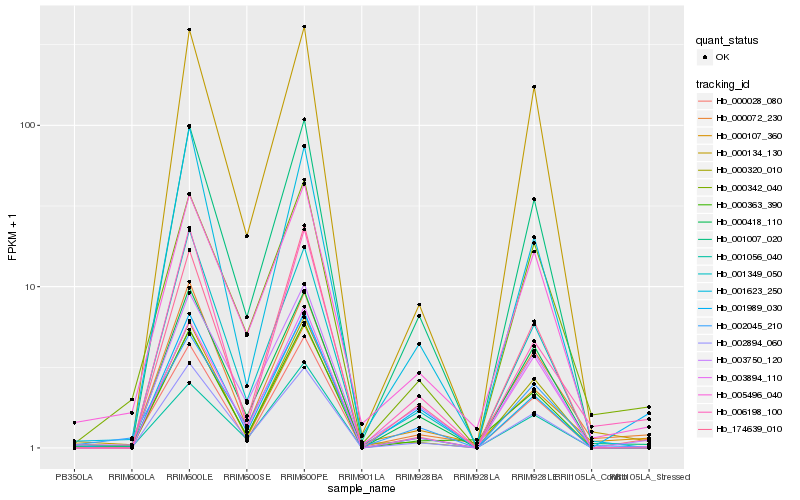
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At3g17950 [Jatropha curcas] |
| 2 |
Hb_001007_020 |
0.1058814772 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000072_230 |
0.1109611332 |
- |
- |
hypothetical protein POPTR_0004s10790g [Populus trichocarpa] |
| 4 |
Hb_174639_010 |
0.1119658818 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_003750_120 |
0.1137926215 |
- |
- |
PREDICTED: uncharacterized protein LOC105112992 [Populus euphratica] |
| 6 |
Hb_006198_100 |
0.1163871033 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 7 |
Hb_000342_040 |
0.1165009997 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 8 |
Hb_000418_110 |
0.1199039422 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 9 |
Hb_002894_060 |
0.1209598049 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 10 |
Hb_001056_040 |
0.1209751965 |
- |
- |
hypothetical protein POPTR_0006s04450g [Populus trichocarpa] |
| 11 |
Hb_003894_110 |
0.1228609495 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 12 |
Hb_000107_360 |
0.1230440244 |
- |
- |
PREDICTED: xylem cysteine proteinase 1 [Jatropha curcas] |
| 13 |
Hb_002045_210 |
0.1235609336 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 14 |
Hb_000363_390 |
0.1278868368 |
- |
- |
Benzoate carboxyl methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_000134_130 |
0.1292670516 |
- |
- |
PREDICTED: uncharacterized protein LOC105628670 [Jatropha curcas] |
| 16 |
Hb_005496_040 |
0.1296580349 |
- |
- |
PREDICTED: uncharacterized protein LOC105630864 [Jatropha curcas] |
| 17 |
Hb_000320_010 |
0.130043874 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001989_030 |
0.1349710728 |
- |
- |
PREDICTED: copper transport protein ATX1 [Jatropha curcas] |
| 19 |
Hb_001349_050 |
0.1359634769 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 20 |
Hb_001623_250 |
0.1365174927 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |