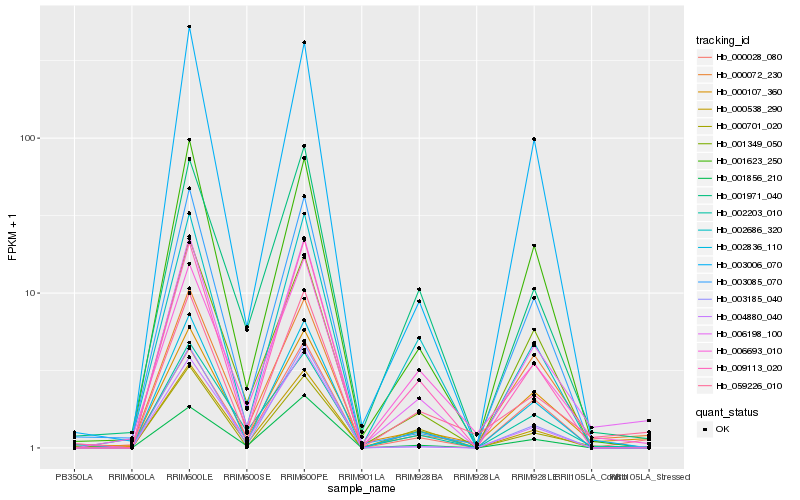
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006198_100 |
0.0 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 2 |
Hb_059226_010 |
0.0862730519 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 3 |
Hb_002203_010 |
0.1051499559 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |
| 4 |
Hb_002836_110 |
0.1052807659 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 5 |
Hb_000538_290 |
0.1067501258 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 6 |
Hb_009113_020 |
0.1102760673 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 7 |
Hb_001623_250 |
0.1114940965 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 8 |
Hb_000107_360 |
0.112297065 |
- |
- |
PREDICTED: xylem cysteine proteinase 1 [Jatropha curcas] |
| 9 |
Hb_000028_080 |
0.1163871033 |
- |
- |
PREDICTED: uncharacterized protein At3g17950 [Jatropha curcas] |
| 10 |
Hb_003006_070 |
0.1185137929 |
- |
- |
aquaporin [Manihot esculenta] |
| 11 |
Hb_000072_230 |
0.1211891439 |
- |
- |
hypothetical protein POPTR_0004s10790g [Populus trichocarpa] |
| 12 |
Hb_001856_210 |
0.1241048212 |
- |
- |
PREDICTED: uncharacterized protein LOC105644171 [Jatropha curcas] |
| 13 |
Hb_004880_040 |
0.1262827893 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001349_050 |
0.1297316227 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 15 |
Hb_002686_320 |
0.1301623523 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 16 |
Hb_000701_020 |
0.1304364916 |
- |
- |
PREDICTED: protein ENDOSPERM DEFECTIVE 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_003185_040 |
0.132486411 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 18 |
Hb_001971_040 |
0.1336684846 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 19 |
Hb_003085_070 |
0.1338885289 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_006693_010 |
0.1348887372 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |