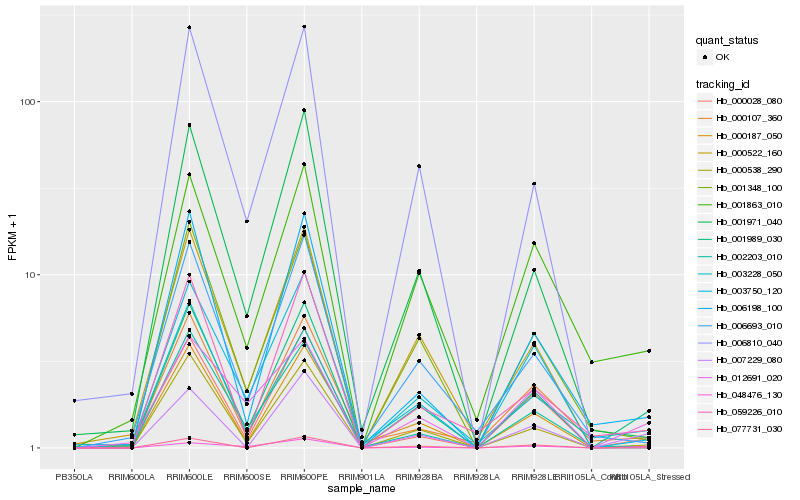
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001989_030 |
0.0 |
- |
- |
PREDICTED: copper transport protein ATX1 [Jatropha curcas] |
| 2 |
Hb_077731_030 |
0.108048545 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 3 |
Hb_007229_080 |
0.1212370496 |
- |
- |
Nonspecific lipid-transfer protein A, putative [Ricinus communis] |
| 4 |
Hb_000028_080 |
0.1349710728 |
- |
- |
PREDICTED: uncharacterized protein At3g17950 [Jatropha curcas] |
| 5 |
Hb_000538_290 |
0.1374741292 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 6 |
Hb_006198_100 |
0.1396089412 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 7 |
Hb_059226_010 |
0.1454456347 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 8 |
Hb_003228_050 |
0.1482964885 |
- |
- |
chromomethylase [Hevea brasiliensis] |
| 9 |
Hb_001348_100 |
0.1504522833 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |
| 10 |
Hb_048476_130 |
0.1563059964 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 11 |
Hb_003750_120 |
0.1599098144 |
- |
- |
PREDICTED: uncharacterized protein LOC105112992 [Populus euphratica] |
| 12 |
Hb_002203_010 |
0.1613153146 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |
| 13 |
Hb_001863_010 |
0.1614111033 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 14 |
Hb_000522_160 |
0.1623788616 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 15 |
Hb_001971_040 |
0.1624759949 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 16 |
Hb_000107_360 |
0.1624812335 |
- |
- |
PREDICTED: xylem cysteine proteinase 1 [Jatropha curcas] |
| 17 |
Hb_006693_010 |
0.1628019294 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |
| 18 |
Hb_012691_020 |
0.1648651138 |
- |
- |
PREDICTED: MFP1 attachment factor 1-like [Jatropha curcas] |
| 19 |
Hb_000187_050 |
0.1658589796 |
- |
- |
PREDICTED: uncharacterized protein LOC105650800 [Jatropha curcas] |
| 20 |
Hb_006810_040 |
0.1662554307 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |