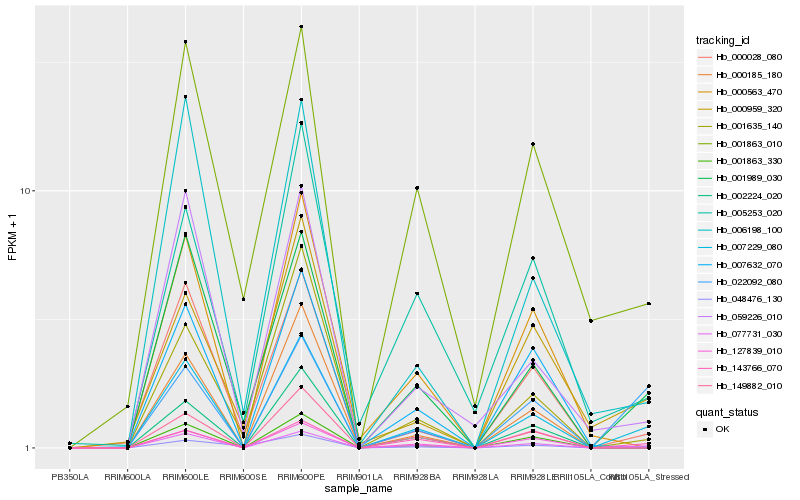
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007229_080 |
0.0 |
- |
- |
Nonspecific lipid-transfer protein A, putative [Ricinus communis] |
| 2 |
Hb_077731_030 |
0.0503266977 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 3 |
Hb_001989_030 |
0.1212370496 |
- |
- |
PREDICTED: copper transport protein ATX1 [Jatropha curcas] |
| 4 |
Hb_000028_080 |
0.1633743078 |
- |
- |
PREDICTED: uncharacterized protein At3g17950 [Jatropha curcas] |
| 5 |
Hb_022092_080 |
0.1717469548 |
- |
- |
- |
| 6 |
Hb_001635_140 |
0.1801488256 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_002224_020 |
0.181303732 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 8 |
Hb_059226_010 |
0.1819561844 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 9 |
Hb_006198_100 |
0.1824945789 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 10 |
Hb_149882_010 |
0.1851776203 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Populus euphratica] |
| 11 |
Hb_001863_330 |
0.1855392617 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000185_180 |
0.188839085 |
- |
- |
hypothetical protein CISIN_1g037968mg, partial [Citrus sinensis] |
| 13 |
Hb_127839_010 |
0.1900479664 |
- |
- |
hypothetical protein PRUPE_ppa000956mg [Prunus persica] |
| 14 |
Hb_000959_320 |
0.1910381418 |
- |
- |
PREDICTED: uncharacterized protein LOC105778325 [Gossypium raimondii] |
| 15 |
Hb_048476_130 |
0.1913195538 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 16 |
Hb_000563_470 |
0.191532925 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 17 |
Hb_143766_070 |
0.1940039056 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 18 |
Hb_005253_020 |
0.196540178 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 19 |
Hb_001863_010 |
0.1989756238 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 20 |
Hb_007632_070 |
0.199106689 |
- |
- |
kinase, putative [Ricinus communis] |