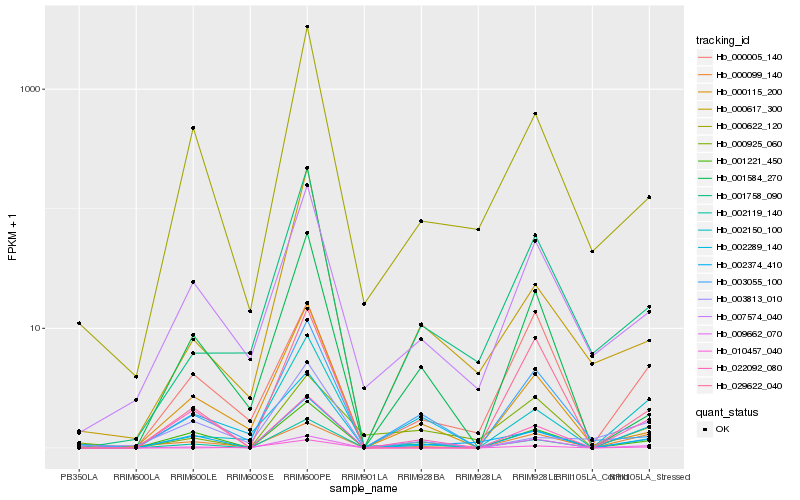
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002119_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634172 [Jatropha curcas] |
| 2 |
Hb_002150_100 |
0.1973098027 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase isoform X1 [Jatropha curcas] |
| 3 |
Hb_001221_450 |
0.2055936064 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000099_140 |
0.2268680625 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 5 |
Hb_002374_410 |
0.2331037426 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_007574_040 |
0.2410259409 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 7 |
Hb_022092_080 |
0.2449351052 |
- |
- |
- |
| 8 |
Hb_001758_090 |
0.2493204024 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 9 |
Hb_003055_100 |
0.2500430538 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 10 |
Hb_000115_200 |
0.253195574 |
- |
- |
PREDICTED: endonuclease 2 [Jatropha curcas] |
| 11 |
Hb_002289_140 |
0.2545849044 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000622_120 |
0.2556730544 |
- |
- |
hypothetical protein [Hevea brasiliensis] |
| 13 |
Hb_001584_270 |
0.257774283 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003813_010 |
0.2621285293 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 1-like [Jatropha curcas] |
| 15 |
Hb_000005_140 |
0.2645224803 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa] |
| 16 |
Hb_000925_060 |
0.2670594361 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 9-like [Jatropha curcas] |
| 17 |
Hb_010457_040 |
0.2678446905 |
- |
- |
PREDICTED: uncharacterized protein LOC101491058 [Cicer arietinum] |
| 18 |
Hb_000617_300 |
0.2747235176 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 19 |
Hb_009662_070 |
0.2748904469 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 20 |
Hb_029622_040 |
0.2752012457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |