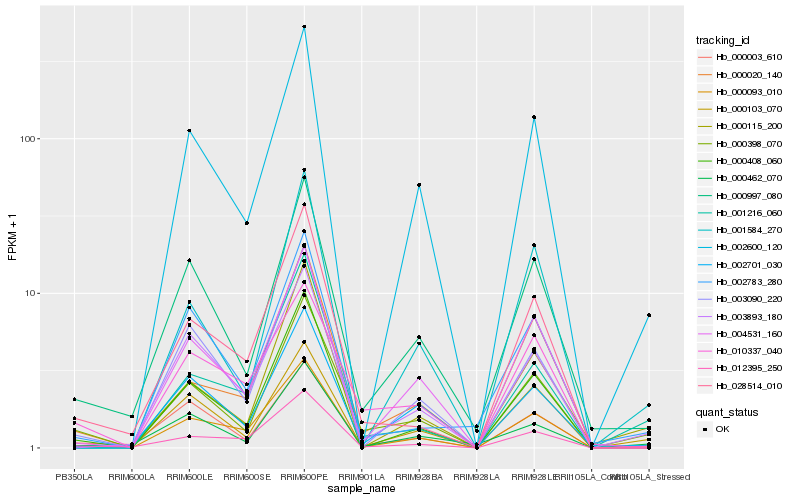
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000398_070 |
0.0 |
- |
- |
hypothetical protein VITISV_011880 [Vitis vinifera] |
| 2 |
Hb_000997_080 |
0.1284955729 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_012395_250 |
0.1417348981 |
- |
- |
hypothetical protein RCOM_1217340 [Ricinus communis] |
| 4 |
Hb_000408_060 |
0.1504594389 |
- |
- |
hypothetical protein POPTR_0018s11360g [Populus trichocarpa] |
| 5 |
Hb_000462_070 |
0.1580613039 |
- |
- |
PREDICTED: probable pectinesterase 15 [Jatropha curcas] |
| 6 |
Hb_000093_010 |
0.1581206438 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 7 |
Hb_002600_120 |
0.1602909395 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 8 |
Hb_000115_200 |
0.1603878863 |
- |
- |
PREDICTED: endonuclease 2 [Jatropha curcas] |
| 9 |
Hb_000020_140 |
0.1622054525 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 10 |
Hb_002701_030 |
0.1628539987 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 11 |
Hb_028514_010 |
0.1640280895 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A9 [Jatropha curcas] |
| 12 |
Hb_004531_160 |
0.1663501411 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000003_610 |
0.1663978776 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase [Jatropha curcas] |
| 14 |
Hb_003893_180 |
0.16672253 |
- |
- |
PREDICTED: uncharacterized protein LOC105648771 [Jatropha curcas] |
| 15 |
Hb_000103_070 |
0.1675478511 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 16 |
Hb_003090_220 |
0.1678153221 |
- |
- |
PREDICTED: copper transporter 4 [Jatropha curcas] |
| 17 |
Hb_001584_270 |
0.1690491209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002783_280 |
0.1698277669 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 19 |
Hb_001216_060 |
0.1725196743 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_010337_040 |
0.1726019897 |
- |
- |
- |