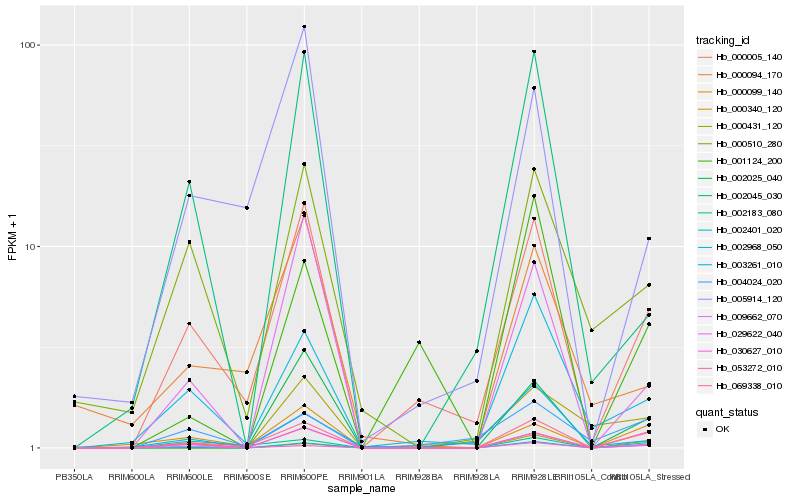
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_069338_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_053272_010 |
0.162252564 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 3 |
Hb_000005_140 |
0.1735542622 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa] |
| 4 |
Hb_009662_070 |
0.2010861259 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 5 |
Hb_002183_080 |
0.2173244821 |
- |
- |
catalytic, putative [Ricinus communis] |
| 6 |
Hb_000099_140 |
0.2180214993 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 7 |
Hb_002968_050 |
0.2341103007 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 8 |
Hb_002401_020 |
0.2389599691 |
- |
- |
PREDICTED: protein argonaute 16 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000510_280 |
0.2613481031 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like [Jatropha curcas] |
| 10 |
Hb_002025_040 |
0.2696803685 |
- |
- |
PREDICTED: WW domain-binding protein 4 [Jatropha curcas] |
| 11 |
Hb_029622_040 |
0.2699953162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004024_020 |
0.271395118 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_030627_010 |
0.271902125 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 [Sesamum indicum] |
| 14 |
Hb_000340_120 |
0.2749293816 |
- |
- |
PREDICTED: uncharacterized protein LOC103406091 [Malus domestica] |
| 15 |
Hb_005914_120 |
0.2775056171 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 16 |
Hb_001124_200 |
0.2944447244 |
- |
- |
hypothetical protein B456_011G186000 [Gossypium raimondii] |
| 17 |
Hb_000431_120 |
0.2959176954 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1-like [Jatropha curcas] |
| 18 |
Hb_000094_170 |
0.2962532145 |
- |
- |
PREDICTED: uncharacterized protein LOC105633781 [Jatropha curcas] |
| 19 |
Hb_003261_010 |
0.2978973604 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 20 |
Hb_002045_030 |
0.3003857508 |
transcription factor |
TF Family: AUX/IAA |
hypothetical protein CISIN_1g047687mg [Citrus sinensis] |