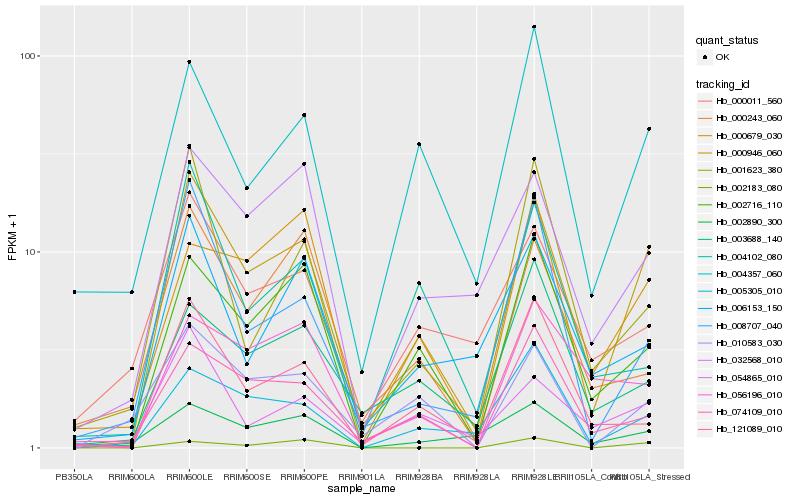
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000946_060 |
0.0 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_002716_110 |
0.1662250101 |
- |
- |
PREDICTED: interaptin [Jatropha curcas] |
| 3 |
Hb_032568_010 |
0.1921921377 |
- |
- |
PREDICTED: protein phosphatase 2C 56-like [Jatropha curcas] |
| 4 |
Hb_005305_010 |
0.193997899 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 5 |
Hb_074109_010 |
0.196444338 |
- |
- |
PREDICTED: putative auxin efflux carrier component 5 [Jatropha curcas] |
| 6 |
Hb_004357_060 |
0.2009366739 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 7 |
Hb_010583_030 |
0.2010172523 |
- |
- |
hypothetical protein PHAVU_011G114500g, partial [Phaseolus vulgaris] |
| 8 |
Hb_008707_040 |
0.2017245575 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_121089_010 |
0.2062801898 |
- |
- |
PREDICTED: uncharacterized protein LOC105645663 [Jatropha curcas] |
| 10 |
Hb_000679_030 |
0.206501688 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5 [Jatropha curcas] |
| 11 |
Hb_054865_010 |
0.2079983007 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 12 |
Hb_001623_380 |
0.2086262609 |
- |
- |
PREDICTED: 7-hydroxymethyl chlorophyll a reductase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002890_300 |
0.2115450915 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 14 |
Hb_006153_150 |
0.2195716806 |
- |
- |
PREDICTED: uncharacterized protein LOC105645999 [Jatropha curcas] |
| 15 |
Hb_004102_080 |
0.2201603391 |
- |
- |
PREDICTED: acyl-protein thioesterase 2 [Jatropha curcas] |
| 16 |
Hb_000243_060 |
0.2225849492 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 17 |
Hb_056196_010 |
0.2241947357 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 18 |
Hb_003688_140 |
0.2244988808 |
- |
- |
PREDICTED: uncharacterized protein LOC105640628 [Jatropha curcas] |
| 19 |
Hb_000011_560 |
0.2253141948 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002183_080 |
0.2268019988 |
- |
- |
catalytic, putative [Ricinus communis] |