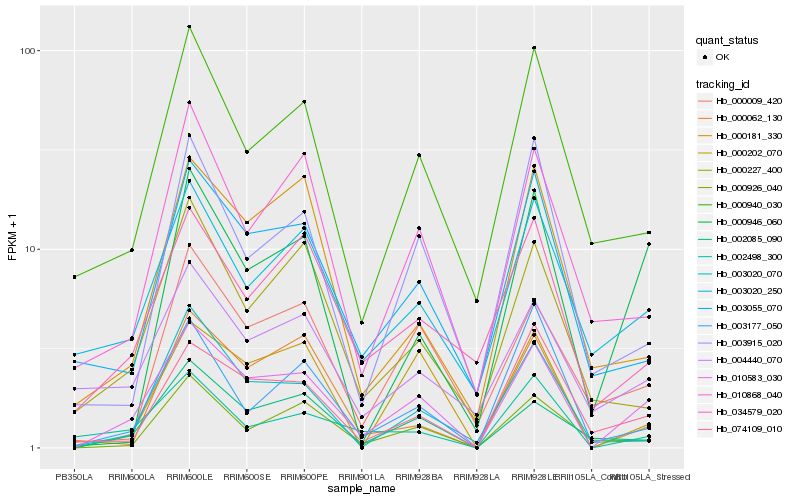
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010583_030 |
0.0 |
- |
- |
hypothetical protein PHAVU_011G114500g, partial [Phaseolus vulgaris] |
| 2 |
Hb_002085_090 |
0.1869722921 |
- |
- |
- |
| 3 |
Hb_000202_070 |
0.189003168 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 4 |
Hb_074109_010 |
0.1940209313 |
- |
- |
PREDICTED: putative auxin efflux carrier component 5 [Jatropha curcas] |
| 5 |
Hb_003055_070 |
0.19703422 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 6 |
Hb_000940_030 |
0.1990664414 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000946_060 |
0.2010172523 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_010868_040 |
0.2022346032 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 9 |
Hb_003020_250 |
0.2023873526 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 10 |
Hb_000009_420 |
0.2032175793 |
- |
- |
PREDICTED: uncharacterized protein LOC105639614 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000926_040 |
0.2036721923 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH5 [Jatropha curcas] |
| 12 |
Hb_034579_020 |
0.2056620959 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002498_300 |
0.2066663435 |
- |
- |
BnaC04g50140D [Brassica napus] |
| 14 |
Hb_003020_070 |
0.2104607494 |
- |
- |
PREDICTED: uncharacterized protein LOC103997323 [Musa acuminata subsp. malaccensis] |
| 15 |
Hb_003915_020 |
0.2112956275 |
- |
- |
PREDICTED: outer envelope pore protein 37, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000062_130 |
0.2126773486 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_000227_400 |
0.2128261341 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 18 |
Hb_004440_070 |
0.2131982943 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520 [Jatropha curcas] |
| 19 |
Hb_000181_330 |
0.2132069601 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 20 |
Hb_003177_050 |
0.2135850089 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Populus euphratica] |