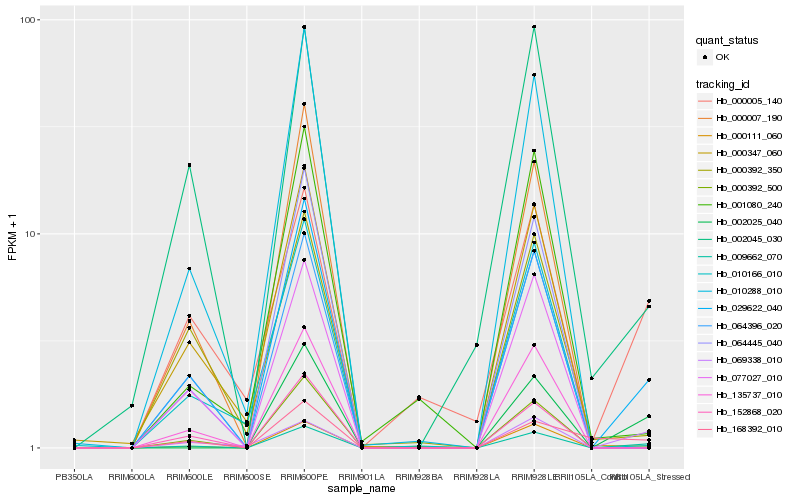
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009662_070 |
0.0 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 2 |
Hb_029622_040 |
0.089790748 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002025_040 |
0.1493275751 |
- |
- |
PREDICTED: WW domain-binding protein 4 [Jatropha curcas] |
| 4 |
Hb_000005_140 |
0.1859958872 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa] |
| 5 |
Hb_069338_010 |
0.2010861259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002045_030 |
0.2042271266 |
transcription factor |
TF Family: AUX/IAA |
hypothetical protein CISIN_1g047687mg [Citrus sinensis] |
| 7 |
Hb_135737_010 |
0.2145921005 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 8 |
Hb_000392_500 |
0.2171203618 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_064445_040 |
0.2201837439 |
- |
- |
hypothetical protein POPTR_0005s09430g [Populus trichocarpa] |
| 10 |
Hb_064396_020 |
0.2202449607 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 11 |
Hb_010288_010 |
0.220624659 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 12 |
Hb_077027_010 |
0.2213766963 |
- |
- |
hypothetical protein JCGZ_04511 [Jatropha curcas] |
| 13 |
Hb_000007_190 |
0.2217819879 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 10-like [Jatropha curcas] |
| 14 |
Hb_000111_060 |
0.2230581904 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 6 isoform X2 [Jatropha curcas] |
| 15 |
Hb_010166_010 |
0.2232263307 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 16 |
Hb_168392_010 |
0.2266378377 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 17 |
Hb_001080_240 |
0.226946456 |
- |
- |
hypothetical protein PHAVU_001G152800g [Phaseolus vulgaris] |
| 18 |
Hb_152868_020 |
0.2271525767 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 19 |
Hb_000392_350 |
0.227178992 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_000347_060 |
0.2276826842 |
- |
- |
PREDICTED: uncharacterized protein LOC105631356 [Jatropha curcas] |