| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
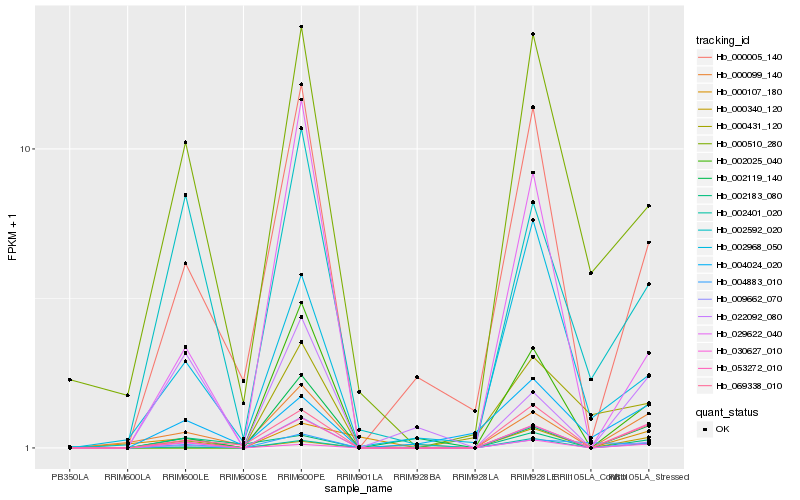
Hb_053272_010 |
0.0 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 2 |
Hb_069338_010 |
0.162252564 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000099_140 |
0.2012109788 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 4 |
Hb_009662_070 |
0.2377888812 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 5 |
Hb_000005_140 |
0.2499470139 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa] |
| 6 |
Hb_002401_020 |
0.253687299 |
- |
- |
PREDICTED: protein argonaute 16 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002025_040 |
0.2820278907 |
- |
- |
PREDICTED: WW domain-binding protein 4 [Jatropha curcas] |
| 8 |
Hb_030627_010 |
0.2855967084 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 [Sesamum indicum] |
| 9 |
Hb_002119_140 |
0.2885928819 |
- |
- |
PREDICTED: uncharacterized protein LOC105634172 [Jatropha curcas] |
| 10 |
Hb_002183_080 |
0.292672799 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_004024_020 |
0.29671849 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000340_120 |
0.3009914954 |
- |
- |
PREDICTED: uncharacterized protein LOC103406091 [Malus domestica] |
| 13 |
Hb_029622_040 |
0.3023014229 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_022092_080 |
0.3074118786 |
- |
- |
- |
| 15 |
Hb_002592_020 |
0.3075975746 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000431_120 |
0.3077151786 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1-like [Jatropha curcas] |
| 17 |
Hb_000510_280 |
0.3104509187 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like [Jatropha curcas] |
| 18 |
Hb_000107_180 |
0.312399039 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103343662 [Prunus mume] |
| 19 |
Hb_002968_050 |
0.3189176523 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 20 |
Hb_004883_010 |
0.3246795629 |
- |
- |
hypothetical protein JCGZ_23144 [Jatropha curcas] |