| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
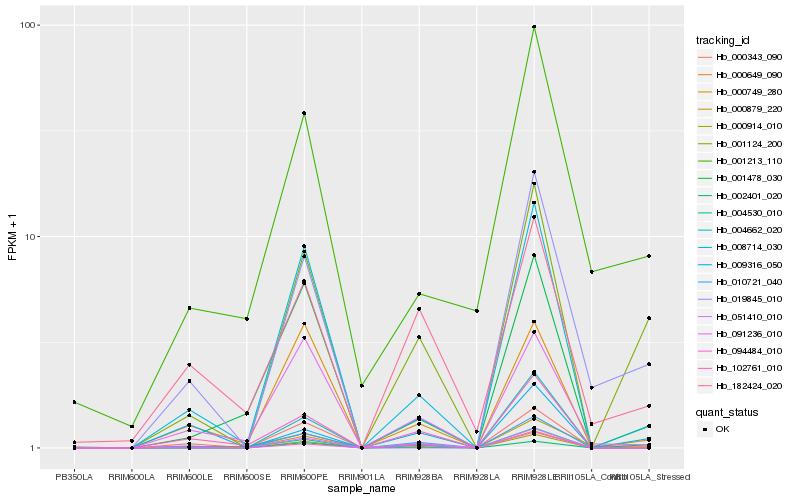
Hb_001124_200 |
0.0 |
- |
- |
hypothetical protein B456_011G186000 [Gossypium raimondii] |
| 2 |
Hb_004662_020 |
0.1535631385 |
- |
- |
40S ribosomal protein S9, putative [Ricinus communis] |
| 3 |
Hb_010721_040 |
0.2359944332 |
transcription factor |
TF Family: M-type |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_002401_020 |
0.2390128954 |
- |
- |
PREDICTED: protein argonaute 16 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001478_030 |
0.2405500777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000879_220 |
0.2462096629 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 7 |
Hb_000749_280 |
0.2495602618 |
- |
- |
PREDICTED: uncharacterized protein LOC105138169 [Populus euphratica] |
| 8 |
Hb_000914_010 |
0.2508887308 |
- |
- |
hypothetical protein POPTR_0025s00760g [Populus trichocarpa] |
| 9 |
Hb_004530_010 |
0.2510245716 |
- |
- |
PREDICTED: uncharacterized protein LOC104898079 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_001213_110 |
0.251341597 |
- |
- |
SOUL heme-binding family protein [Populus trichocarpa] |
| 11 |
Hb_051410_010 |
0.2517584467 |
- |
- |
PREDICTED: uncharacterized protein LOC103401813 [Malus domestica] |
| 12 |
Hb_182424_020 |
0.2557334267 |
- |
- |
hypothetical protein POPTR_0006s24190g [Populus trichocarpa] |
| 13 |
Hb_008714_030 |
0.2571005136 |
- |
- |
Multidrug resistance protein ABC transporter family protein, putative [Theobroma cacao] |
| 14 |
Hb_000343_090 |
0.2580216468 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_019845_010 |
0.2591515472 |
- |
- |
protease inhibitor protein [Hevea brasiliensis] |
| 16 |
Hb_000649_090 |
0.2626535032 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_094484_010 |
0.2640785804 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_102761_010 |
0.2644220561 |
- |
- |
hypothetical protein JCGZ_21614 [Jatropha curcas] |
| 19 |
Hb_091236_010 |
0.267381143 |
- |
- |
hypothetical protein POPTR_0019s12290g [Populus trichocarpa] |
| 20 |
Hb_009316_050 |
0.2677489568 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |