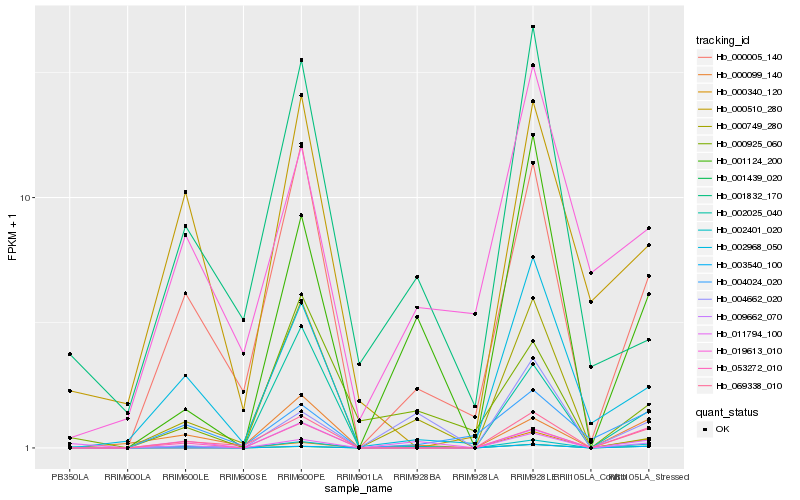
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002401_020 |
0.0 |
- |
- |
PREDICTED: protein argonaute 16 isoform X2 [Jatropha curcas] |
| 2 |
Hb_069338_010 |
0.2389599691 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001124_200 |
0.2390128954 |
- |
- |
hypothetical protein B456_011G186000 [Gossypium raimondii] |
| 4 |
Hb_011794_100 |
0.24502227 |
- |
- |
PREDICTED: seven transmembrane domain-containing tyrosine-protein kinase 1-like [Jatropha curcas] |
| 5 |
Hb_000005_140 |
0.2507853617 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa] |
| 6 |
Hb_053272_010 |
0.253687299 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 7 |
Hb_009662_070 |
0.2755023931 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 8 |
Hb_000099_140 |
0.2851795412 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 9 |
Hb_002968_050 |
0.2884447988 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 10 |
Hb_003540_100 |
0.2908528282 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 11 |
Hb_000925_060 |
0.2918150447 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 9-like [Jatropha curcas] |
| 12 |
Hb_004024_020 |
0.3045418249 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000340_120 |
0.3060271042 |
- |
- |
PREDICTED: uncharacterized protein LOC103406091 [Malus domestica] |
| 14 |
Hb_001439_020 |
0.3069822664 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 15 |
Hb_001832_170 |
0.3148890949 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 16 |
Hb_000510_280 |
0.3166409245 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like [Jatropha curcas] |
| 17 |
Hb_002025_040 |
0.3191789999 |
- |
- |
PREDICTED: WW domain-binding protein 4 [Jatropha curcas] |
| 18 |
Hb_000749_280 |
0.3196490026 |
- |
- |
PREDICTED: uncharacterized protein LOC105138169 [Populus euphratica] |
| 19 |
Hb_004662_020 |
0.3254636946 |
- |
- |
40S ribosomal protein S9, putative [Ricinus communis] |
| 20 |
Hb_019613_010 |
0.3257060738 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |