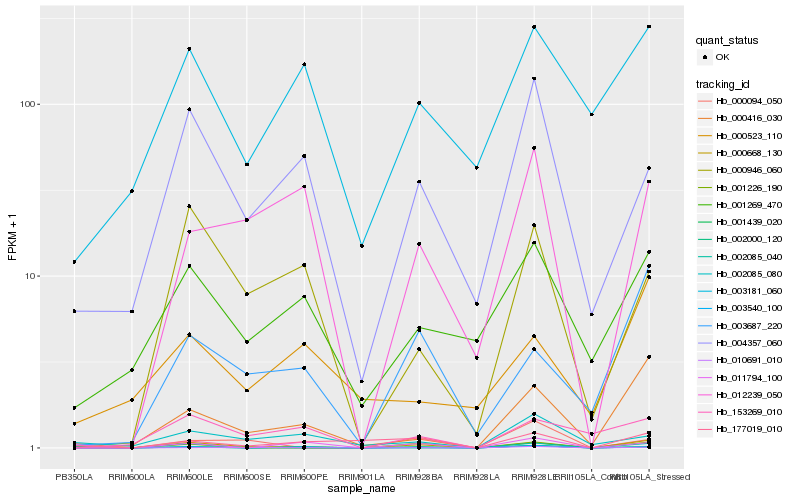
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000668_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_003687_220 |
0.2670350703 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 3 |
Hb_177019_010 |
0.2922440685 |
- |
- |
Uncharacterized protein TCM_004012 [Theobroma cacao] |
| 4 |
Hb_011794_100 |
0.2950891203 |
- |
- |
PREDICTED: seven transmembrane domain-containing tyrosine-protein kinase 1-like [Jatropha curcas] |
| 5 |
Hb_000416_030 |
0.2951284106 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001226_190 |
0.3037886758 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 7 |
Hb_002000_120 |
0.3077792991 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_002085_040 |
0.3122012926 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 9 |
Hb_004357_060 |
0.31488451 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 10 |
Hb_010691_010 |
0.3164535388 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 11 |
Hb_001269_470 |
0.3206882514 |
- |
- |
PREDICTED: uncharacterized protein LOC105630313 [Jatropha curcas] |
| 12 |
Hb_001439_020 |
0.3289819026 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 13 |
Hb_000094_050 |
0.3292690075 |
- |
- |
- |
| 14 |
Hb_003181_060 |
0.3324131854 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Jatropha curcas] |
| 15 |
Hb_000946_060 |
0.3342401291 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_153269_010 |
0.3355214472 |
- |
- |
hypothetical protein CICLE_v10029937mg [Citrus clementina] |
| 17 |
Hb_002085_080 |
0.3364964683 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 68-like [Jatropha curcas] |
| 18 |
Hb_012239_050 |
0.3379871747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003540_100 |
0.3381509242 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 20 |
Hb_000523_110 |
0.3403354109 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |