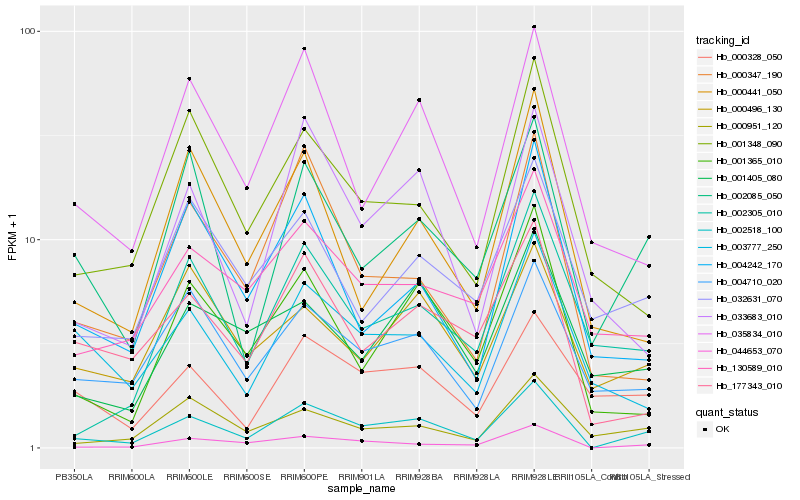
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002518_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_044653_070 |
0.1474168122 |
- |
- |
hypothetical protein CICLE_v10018540mg [Citrus clementina] |
| 3 |
Hb_002085_050 |
0.1661851193 |
- |
- |
PREDICTED: beta-galactosidase 8 [Jatropha curcas] |
| 4 |
Hb_001365_010 |
0.1784913896 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 5 |
Hb_035834_010 |
0.1787863954 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_177343_010 |
0.180538751 |
- |
- |
PREDICTED: uncharacterized protein LOC105650963 [Jatropha curcas] |
| 7 |
Hb_004710_020 |
0.1807185552 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 8 |
Hb_033683_010 |
0.1868640951 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 9 |
Hb_130589_010 |
0.188553185 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 10 |
Hb_000951_120 |
0.1899519055 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 11 |
Hb_000347_190 |
0.1899606501 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 12 |
Hb_000328_050 |
0.1948581985 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001348_090 |
0.1952125722 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000496_130 |
0.1963847339 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 15 |
Hb_004242_170 |
0.1964167126 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 16 |
Hb_032631_070 |
0.1974124544 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 17 |
Hb_001405_080 |
0.1975545117 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 18 |
Hb_003777_250 |
0.1983419198 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 19 |
Hb_000441_050 |
0.1984004945 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 20 |
Hb_002305_010 |
0.1985775576 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |