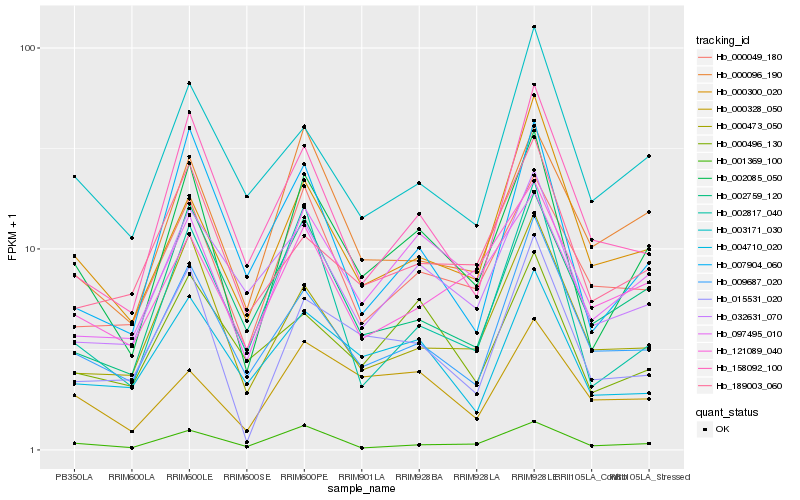
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002085_050 |
0.0 |
- |
- |
PREDICTED: beta-galactosidase 8 [Jatropha curcas] |
| 2 |
Hb_097495_010 |
0.1417255575 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 3 |
Hb_015531_020 |
0.1429705201 |
- |
- |
PREDICTED: uncharacterized protein LOC100267182 isoform X2 [Vitis vinifera] |
| 4 |
Hb_009687_020 |
0.144407195 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 5 |
Hb_001369_100 |
0.1455781246 |
- |
- |
hypothetical protein JCGZ_26280 [Jatropha curcas] |
| 6 |
Hb_002759_120 |
0.1483547183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000049_180 |
0.1488223504 |
- |
- |
aromatic amino acid decarboxylase, putative [Ricinus communis] |
| 8 |
Hb_000096_190 |
0.1514114362 |
- |
- |
unknown [Lotus japonicus] |
| 9 |
Hb_000473_050 |
0.151513512 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 10 |
Hb_007904_060 |
0.1523612944 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1 [Jatropha curcas] |
| 11 |
Hb_158092_100 |
0.153527641 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 12 |
Hb_032631_070 |
0.1548707294 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_002817_040 |
0.1568964215 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 14 |
Hb_004710_020 |
0.1580964505 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 15 |
Hb_000300_020 |
0.1591946181 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 16 |
Hb_003171_030 |
0.1619858106 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000496_130 |
0.1623754084 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 18 |
Hb_121089_040 |
0.1633301261 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 19 |
Hb_000328_050 |
0.1637213542 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_189003_060 |
0.1646093488 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |