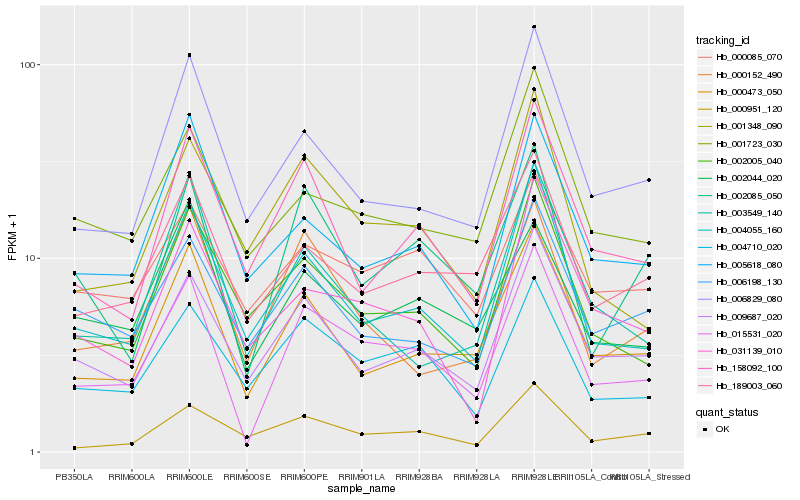
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015531_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100267182 isoform X2 [Vitis vinifera] |
| 2 |
Hb_000473_050 |
0.1368040773 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 3 |
Hb_004055_160 |
0.13806519 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_009687_020 |
0.1415694154 |
- |
- |
hypothetical protein JCGZ_10323 [Jatropha curcas] |
| 5 |
Hb_002085_050 |
0.1429705201 |
- |
- |
PREDICTED: beta-galactosidase 8 [Jatropha curcas] |
| 6 |
Hb_004710_020 |
0.1442753374 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 7 |
Hb_001348_090 |
0.1538717701 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 8 |
Hb_006829_080 |
0.1560999815 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 9 |
Hb_189003_060 |
0.15616234 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_002005_040 |
0.1564067735 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_158092_100 |
0.1588039188 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 12 |
Hb_000152_490 |
0.1608444502 |
- |
- |
PREDICTED: (6-4)DNA photolyase [Jatropha curcas] |
| 13 |
Hb_001723_030 |
0.1609558491 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002044_020 |
0.1627957537 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 15 |
Hb_000951_120 |
0.1627998292 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 16 |
Hb_000085_070 |
0.1633132875 |
- |
- |
PREDICTED: trigger factor-like protein TIG, Chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_006198_130 |
0.1665665857 |
- |
- |
PREDICTED: uncharacterized protein LOC105644406 [Jatropha curcas] |
| 18 |
Hb_031139_010 |
0.1666951491 |
- |
- |
PREDICTED: uncharacterized protein LOC105632403 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003549_140 |
0.1699560449 |
- |
- |
Thylakoid lumenal 21.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_005618_080 |
0.1701353339 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |