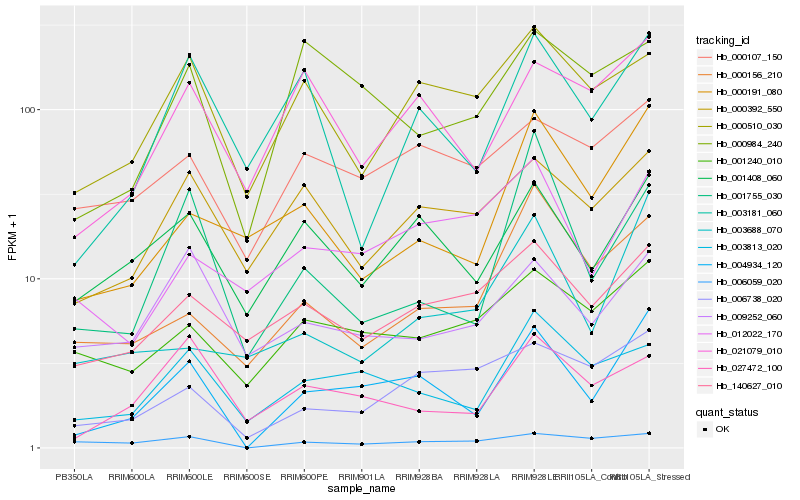
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004934_120 |
0.0 |
- |
- |
- |
| 2 |
Hb_001408_060 |
0.2003126235 |
- |
- |
PREDICTED: 60S ribosomal protein L3 [Eucalyptus grandis] |
| 3 |
Hb_003813_020 |
0.210478696 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 4 |
Hb_021079_010 |
0.2161125156 |
- |
- |
PREDICTED: histone H2B-like [Populus euphratica] |
| 5 |
Hb_000191_080 |
0.2232837763 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 6 |
Hb_006738_020 |
0.2255048574 |
- |
- |
hypothetical protein VITISV_010510 [Vitis vinifera] |
| 7 |
Hb_001240_010 |
0.2295687845 |
- |
- |
PREDICTED: uncharacterized protein LOC105634101 [Jatropha curcas] |
| 8 |
Hb_003181_060 |
0.231190294 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Jatropha curcas] |
| 9 |
Hb_000107_150 |
0.2318686287 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 10 |
Hb_000392_550 |
0.2328386502 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 6, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_140627_010 |
0.2350404176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_027472_100 |
0.2371842678 |
- |
- |
PREDICTED: uncharacterized protein LOC105646173 [Jatropha curcas] |
| 13 |
Hb_003688_070 |
0.2373866101 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein E [Jatropha curcas] |
| 14 |
Hb_012022_170 |
0.2385778708 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 15 |
Hb_001755_030 |
0.2396521072 |
- |
- |
PREDICTED: D-glycerate 3-kinase, chloroplastic-like [Populus euphratica] |
| 16 |
Hb_000984_240 |
0.2397572872 |
- |
- |
hypothetical protein JCGZ_15962 [Jatropha curcas] |
| 17 |
Hb_000510_030 |
0.2406512718 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 18 |
Hb_009252_060 |
0.2408346085 |
- |
- |
PREDICTED: uncharacterized protein LOC105640395 [Jatropha curcas] |
| 19 |
Hb_000156_210 |
0.2415433898 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 20 |
Hb_006059_020 |
0.2416606869 |
- |
- |
PREDICTED: uncharacterized protein LOC105648672 [Jatropha curcas] |