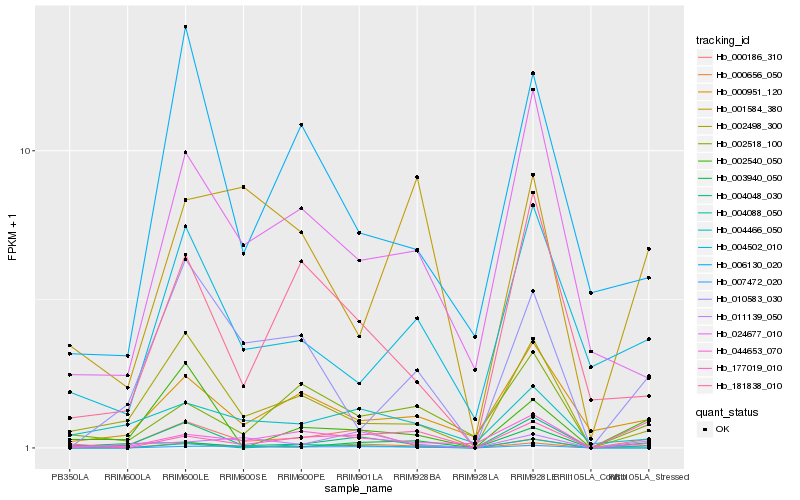
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000656_050 |
0.0 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase II, chloroplastic-like [Vitis vinifera] |
| 2 |
Hb_177019_010 |
0.2750973388 |
- |
- |
Uncharacterized protein TCM_004012 [Theobroma cacao] |
| 3 |
Hb_000186_310 |
0.2885689287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011139_050 |
0.2914035741 |
- |
- |
nicotinate phosphoribosyltransferase family protein [Populus trichocarpa] |
| 5 |
Hb_004088_050 |
0.2957249006 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 6 |
Hb_007472_020 |
0.3182279431 |
- |
- |
hypothetical protein PRUPE_ppa016115mg [Prunus persica] |
| 7 |
Hb_004502_010 |
0.3185446561 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |
| 8 |
Hb_000951_120 |
0.3216892378 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 9 |
Hb_044653_070 |
0.3238435016 |
- |
- |
hypothetical protein CICLE_v10018540mg [Citrus clementina] |
| 10 |
Hb_004048_030 |
0.32704325 |
- |
- |
hypothetical protein B456_009G371200, partial [Gossypium raimondii] |
| 11 |
Hb_004466_050 |
0.3295362181 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |
| 12 |
Hb_024677_010 |
0.3297955466 |
- |
- |
superoxide dismutase [fe], putative [Ricinus communis] |
| 13 |
Hb_002540_050 |
0.3326332342 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g13770, mitochondrial [Jatropha curcas] |
| 14 |
Hb_010583_030 |
0.3341049499 |
- |
- |
hypothetical protein PHAVU_011G114500g, partial [Phaseolus vulgaris] |
| 15 |
Hb_006130_020 |
0.3360499821 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_003940_050 |
0.3363453453 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 17 |
Hb_002498_300 |
0.3366109928 |
- |
- |
BnaC04g50140D [Brassica napus] |
| 18 |
Hb_181838_010 |
0.3382149676 |
- |
- |
PREDICTED: uncharacterized protein LOC105631275 [Jatropha curcas] |
| 19 |
Hb_002518_100 |
0.3400626153 |
- |
- |
- |
| 20 |
Hb_001584_380 |
0.3405266728 |
- |
- |
PREDICTED: tropomyosin-like [Jatropha curcas] |