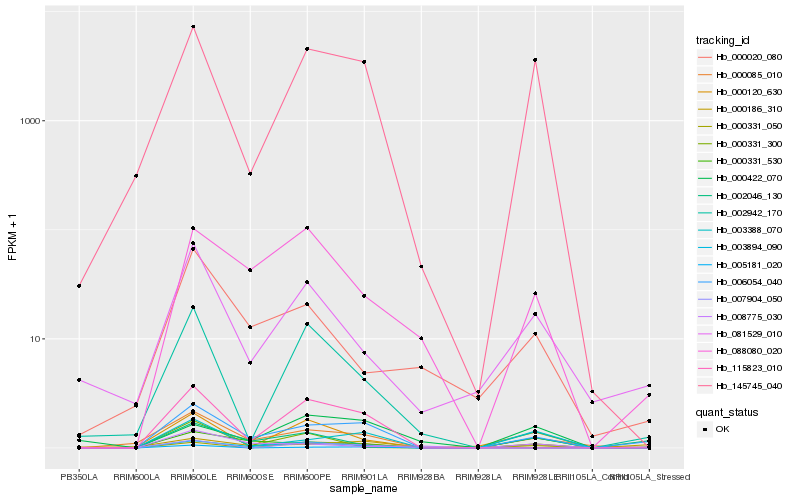
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006054_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_000085_010 |
0.2405673144 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000186_310 |
0.2427354112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_145745_040 |
0.2553293475 |
- |
- |
Cell wall-associated hydrolase [Medicago truncatula] |
| 5 |
Hb_003388_070 |
0.2583136221 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 6 |
Hb_088080_020 |
0.2780813089 |
- |
- |
- |
| 7 |
Hb_115823_010 |
0.2815833848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000331_050 |
0.290152581 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 9 |
Hb_081529_010 |
0.2925971129 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_002046_130 |
0.2962803094 |
- |
- |
PREDICTED: UDP-sugar transporter sqv-7-like [Jatropha curcas] |
| 11 |
Hb_003894_090 |
0.2989652425 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000331_530 |
0.3081528675 |
- |
- |
PREDICTED: callose synthase 10 isoform X1 [Jatropha curcas] |
| 13 |
Hb_008775_030 |
0.3119843511 |
- |
- |
PREDICTED: ferric reduction oxidase 7, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_000120_630 |
0.3163868526 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 15 |
Hb_000020_080 |
0.3184912208 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_09080 [Jatropha curcas] |
| 16 |
Hb_000422_070 |
0.3220427913 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005181_020 |
0.3232578873 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 18 |
Hb_000331_300 |
0.3238494772 |
- |
- |
PREDICTED: malate synthase, glyoxysomal [Jatropha curcas] |
| 19 |
Hb_002942_170 |
0.3263801985 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 20 |
Hb_007904_050 |
0.3276345266 |
- |
- |
hypothetical protein JCGZ_17883 [Jatropha curcas] |