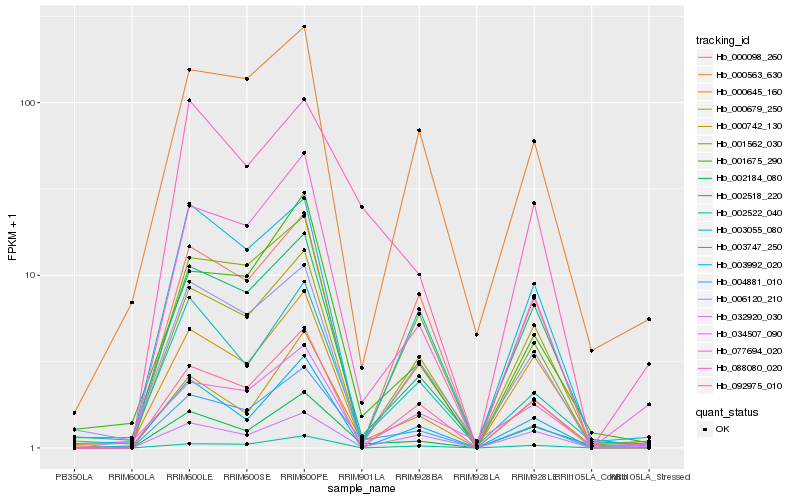
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004881_010 |
0.0 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL1 [Jatropha curcas] |
| 2 |
Hb_077694_020 |
0.0843292211 |
- |
- |
SAUR family protein [Theobroma cacao] |
| 3 |
Hb_000679_250 |
0.1394462981 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000742_130 |
0.1406120901 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g30570 [Jatropha curcas] |
| 5 |
Hb_092975_010 |
0.1505905997 |
- |
- |
hypothetical protein JCGZ_04359 [Jatropha curcas] |
| 6 |
Hb_002184_080 |
0.1519071176 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 7 |
Hb_001675_290 |
0.1526004686 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003992_020 |
0.1534938646 |
- |
- |
PREDICTED: phosphatidylcholine:diacylglycerol cholinephosphotransferase 1-like [Jatropha curcas] |
| 9 |
Hb_000563_630 |
0.1551009673 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 10 |
Hb_001562_030 |
0.1551235561 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 11 |
Hb_034507_090 |
0.1595894306 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002518_220 |
0.1605585028 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 13 |
Hb_088080_020 |
0.1612830946 |
- |
- |
- |
| 14 |
Hb_000098_260 |
0.1618895725 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 15 |
Hb_032920_030 |
0.1624318273 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 16 |
Hb_006120_210 |
0.162839966 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 17 |
Hb_003747_250 |
0.1628800292 |
- |
- |
PREDICTED: potassium transporter 7 isoform X2 [Elaeis guineensis] |
| 18 |
Hb_002522_040 |
0.1642189848 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 19 |
Hb_003055_080 |
0.1644928127 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000645_160 |
0.1653048626 |
- |
- |
protein translocase, putative [Ricinus communis] |