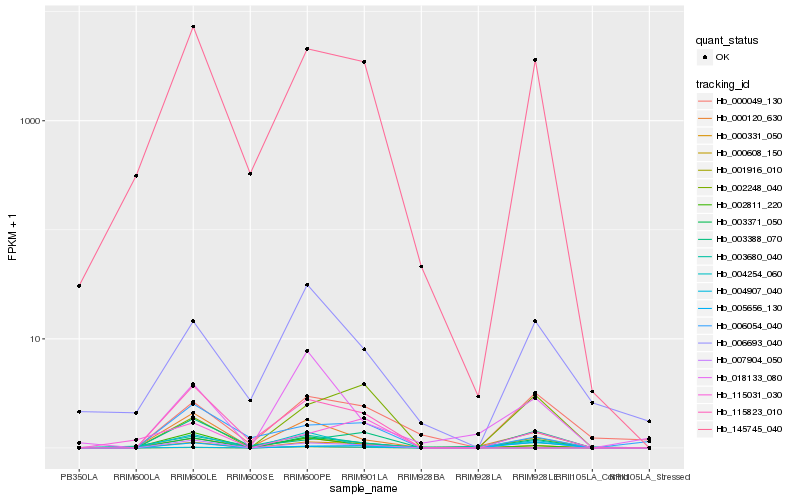
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_050 |
0.0 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 2 |
Hb_003371_050 |
0.1559563845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_145745_040 |
0.1818045662 |
- |
- |
Cell wall-associated hydrolase [Medicago truncatula] |
| 4 |
Hb_000120_630 |
0.2324370283 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 5 |
Hb_002811_220 |
0.2737921373 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_115031_030 |
0.2754896387 |
- |
- |
ribosomal protein S12 (chloroplast) [Camellia sinensis] |
| 7 |
Hb_002248_040 |
0.2774698964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003388_070 |
0.2781459671 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 9 |
Hb_004907_040 |
0.2824420016 |
- |
- |
hypothetical protein POPTR_0011s079751g, partial [Populus trichocarpa] |
| 10 |
Hb_007904_050 |
0.2830489596 |
- |
- |
hypothetical protein JCGZ_17883 [Jatropha curcas] |
| 11 |
Hb_006054_040 |
0.290152581 |
- |
- |
- |
| 12 |
Hb_000608_150 |
0.2920314831 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004254_060 |
0.2922306946 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 14 |
Hb_000049_130 |
0.2951536858 |
- |
- |
PREDICTED: uncharacterized protein LOC104879855 [Vitis vinifera] |
| 15 |
Hb_115823_010 |
0.2992996505 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006693_040 |
0.3002137354 |
- |
- |
PREDICTED: homeotic protein female sterile-like [Jatropha curcas] |
| 17 |
Hb_018133_080 |
0.3104984032 |
- |
- |
- |
| 18 |
Hb_003680_040 |
0.3129350661 |
- |
- |
PREDICTED: polyadenylate-binding protein 7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001916_010 |
0.3134733753 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 20 |
Hb_005656_130 |
0.3156495746 |
- |
- |
PREDICTED: calmodulin-like protein 7 [Jatropha curcas] |