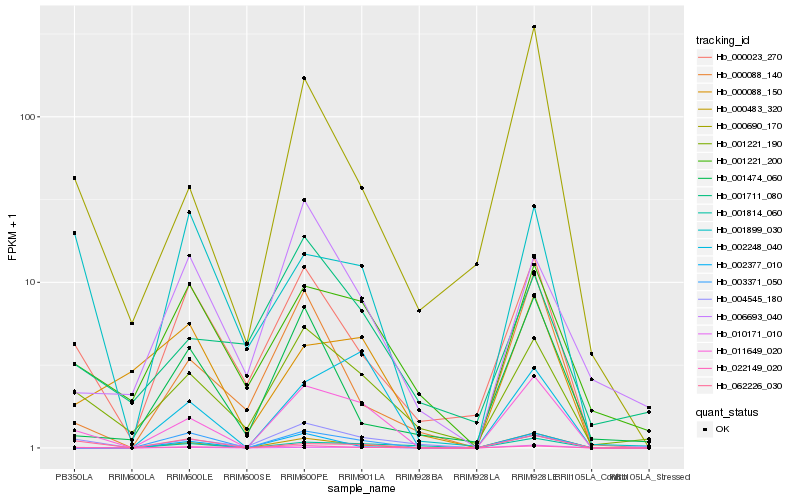
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011649_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104212904 isoform X1 [Nicotiana sylvestris] |
| 2 |
Hb_000483_320 |
0.2267717638 |
- |
- |
Major pollen allergen Ory s 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_001221_190 |
0.2276789437 |
- |
- |
PREDICTED: F-box protein At5g50450 [Jatropha curcas] |
| 4 |
Hb_002248_040 |
0.2569787416 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000690_170 |
0.2571099732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000023_270 |
0.2583851532 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003371_050 |
0.2671208757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001221_200 |
0.2685662265 |
- |
- |
- |
| 9 |
Hb_000088_140 |
0.2699404145 |
- |
- |
- |
| 10 |
Hb_004545_180 |
0.2778145662 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_010171_010 |
0.2778273332 |
- |
- |
hypothetical protein VITISV_033646 [Vitis vinifera] |
| 12 |
Hb_001814_060 |
0.2793207385 |
- |
- |
PREDICTED: uncharacterized protein LOC105650637 [Jatropha curcas] |
| 13 |
Hb_022149_020 |
0.2899961992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000088_150 |
0.2918359769 |
- |
- |
hypothetical protein POPTR_0013s11250g [Populus trichocarpa] |
| 15 |
Hb_002377_010 |
0.2948189641 |
transcription factor |
TF Family: C2C2-CO-like |
PREDICTED: zinc finger protein CONSTANS-LIKE 2-like [Jatropha curcas] |
| 16 |
Hb_062226_030 |
0.2965941211 |
- |
- |
- |
| 17 |
Hb_001899_030 |
0.3046486033 |
- |
- |
hypothetical protein JCGZ_19903 [Jatropha curcas] |
| 18 |
Hb_001711_080 |
0.3054895519 |
transcription factor |
TF Family: C2C2-LSD |
PREDICTED: protein LSD1 isoform X1 [Phoenix dactylifera] |
| 19 |
Hb_001474_060 |
0.30762063 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 20 |
Hb_006693_040 |
0.3100782196 |
- |
- |
PREDICTED: homeotic protein female sterile-like [Jatropha curcas] |