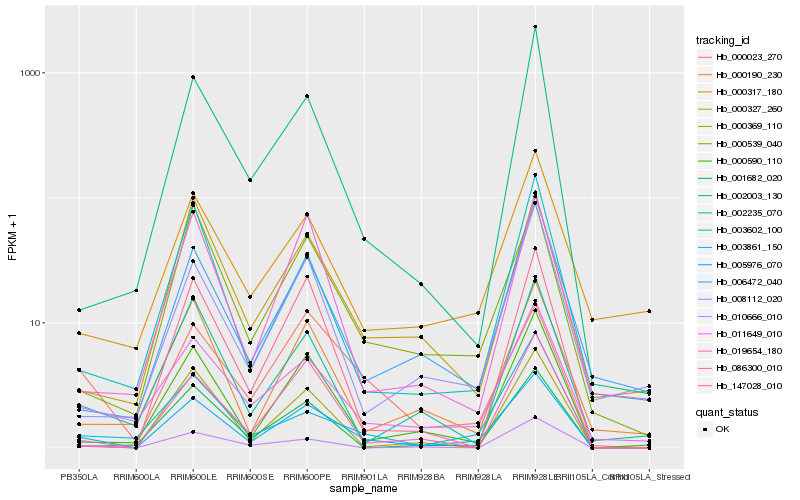
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005976_070 |
0.0 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 2 |
Hb_011649_010 |
0.1530706742 |
transcription factor |
TF Family: Pseudo ARR-B |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 3 |
Hb_000590_110 |
0.1591300393 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000539_040 |
0.1611933911 |
- |
- |
PREDICTED: uncharacterized protein LOC105636110 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002003_130 |
0.1748845607 |
- |
- |
hypothetical protein CISIN_1g027843mg [Citrus sinensis] |
| 6 |
Hb_006472_040 |
0.1799389451 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003602_100 |
0.1800414282 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_000023_270 |
0.1828237969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000190_230 |
0.183295003 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 10 |
Hb_003861_150 |
0.1833314403 |
- |
- |
hypothetical protein JCGZ_15167 [Jatropha curcas] |
| 11 |
Hb_001682_020 |
0.1840821758 |
- |
- |
PREDICTED: uncharacterized protein LOC105628366 [Jatropha curcas] |
| 12 |
Hb_010666_010 |
0.1843723973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000369_110 |
0.1847009623 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_147028_010 |
0.185877495 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Prunus mume] |
| 15 |
Hb_000327_260 |
0.1859860424 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_019654_180 |
0.1865407157 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 17 |
Hb_008112_020 |
0.1868165088 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002235_070 |
0.1873850048 |
- |
- |
PREDICTED: uncharacterized protein LOC105644108 [Jatropha curcas] |
| 19 |
Hb_086300_010 |
0.1887578584 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 20 |
Hb_000317_180 |
0.1900731699 |
- |
- |
PREDICTED: protein THYLAKOID FORMATION1, chloroplastic [Jatropha curcas] |