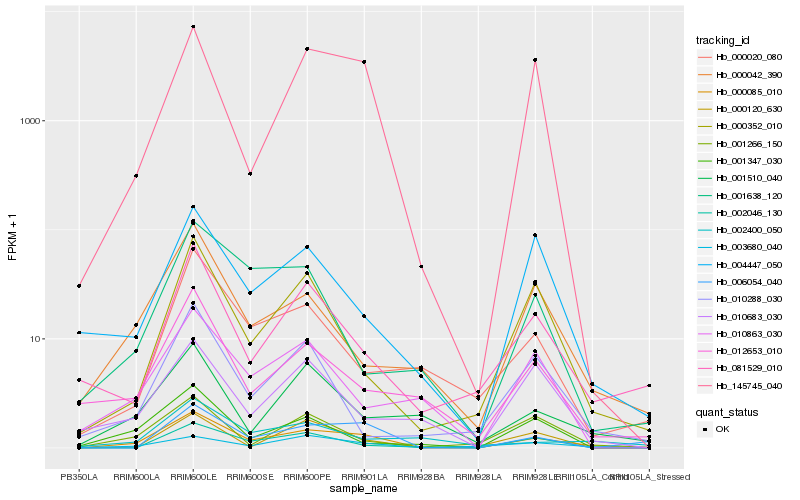
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000085_010 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_145745_040 |
0.2181378583 |
- |
- |
Cell wall-associated hydrolase [Medicago truncatula] |
| 3 |
Hb_006054_040 |
0.2405673144 |
- |
- |
- |
| 4 |
Hb_003680_040 |
0.2412558036 |
- |
- |
PREDICTED: polyadenylate-binding protein 7 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000352_010 |
0.2483161088 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 6 |
Hb_081529_010 |
0.2488941345 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_012653_010 |
0.2494694213 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 8 |
Hb_000042_390 |
0.2498331782 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7 [Jatropha curcas] |
| 9 |
Hb_004447_050 |
0.252827584 |
- |
- |
PREDICTED: high-light-induced protein, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002046_130 |
0.2557691383 |
- |
- |
PREDICTED: UDP-sugar transporter sqv-7-like [Jatropha curcas] |
| 11 |
Hb_010863_030 |
0.2591575923 |
- |
- |
PREDICTED: uncharacterized protein LOC105642466 [Jatropha curcas] |
| 12 |
Hb_001510_040 |
0.2619895111 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein [Jatropha curcas] |
| 13 |
Hb_000020_080 |
0.2632452506 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_09080 [Jatropha curcas] |
| 14 |
Hb_001347_030 |
0.2642864731 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_001266_150 |
0.2691875017 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 16 |
Hb_010683_030 |
0.2710568765 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000120_630 |
0.2721400275 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 18 |
Hb_001638_120 |
0.2723147127 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 19 |
Hb_010288_030 |
0.2726154824 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 20 |
Hb_002400_050 |
0.2765954616 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |