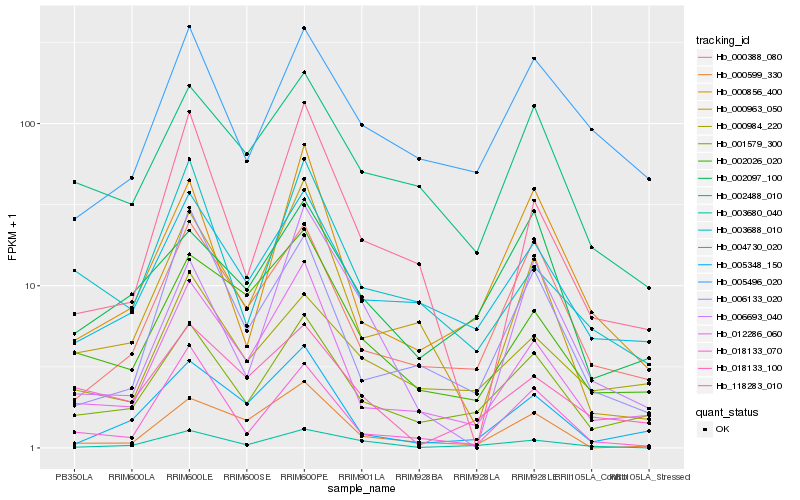
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003680_040 |
0.0 |
- |
- |
PREDICTED: polyadenylate-binding protein 7 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001579_300 |
0.1829203 |
- |
- |
PREDICTED: probable polygalacturonase At1g80170 [Jatropha curcas] |
| 3 |
Hb_004730_020 |
0.1959324111 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 4 |
Hb_005496_020 |
0.1994811503 |
- |
- |
PREDICTED: uncharacterized protein LOC105630867 [Jatropha curcas] |
| 5 |
Hb_000388_080 |
0.2044283765 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 6 |
Hb_000856_400 |
0.2140890748 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 7 |
Hb_000984_220 |
0.2148764259 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 8 |
Hb_002026_020 |
0.2150919282 |
- |
- |
hypothetical protein POPTR_0012s129501g, partial [Populus trichocarpa] |
| 9 |
Hb_012286_060 |
0.2191010162 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_018133_100 |
0.2197564424 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_002488_010 |
0.2202076382 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 12 |
Hb_006133_020 |
0.2203356581 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 13 |
Hb_118283_010 |
0.220555675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006693_040 |
0.2218016644 |
- |
- |
PREDICTED: homeotic protein female sterile-like [Jatropha curcas] |
| 15 |
Hb_003688_010 |
0.2232903384 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3-like [Jatropha curcas] |
| 16 |
Hb_018133_070 |
0.2248543477 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 17 |
Hb_000599_330 |
0.2270601203 |
- |
- |
hypothetical protein JCGZ_17661 [Jatropha curcas] |
| 18 |
Hb_005348_150 |
0.2290577806 |
- |
- |
PREDICTED: actin-100-like [Jatropha curcas] |
| 19 |
Hb_002097_100 |
0.2290737239 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_000963_050 |
0.2292069848 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Jatropha curcas] |