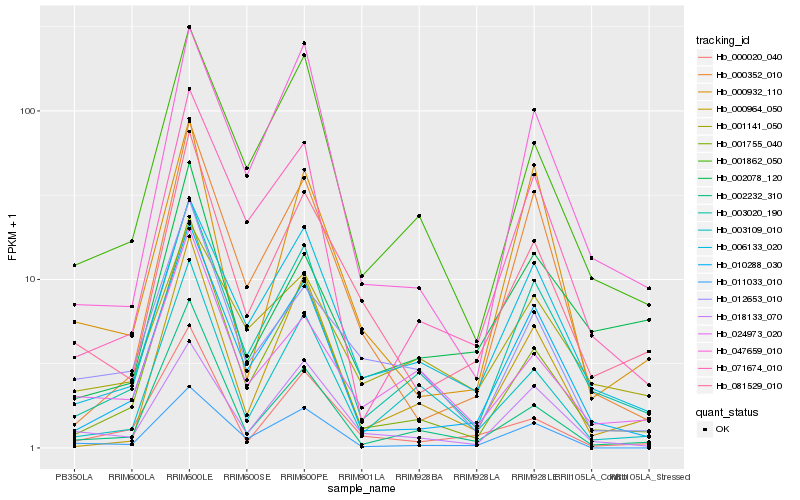
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_081529_010 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_018133_070 |
0.1506191442 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 3 |
Hb_010288_030 |
0.1527243145 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 4 |
Hb_006133_020 |
0.1543563807 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 5 |
Hb_012653_010 |
0.1545610631 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 6 |
Hb_000020_040 |
0.1552899652 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_003020_190 |
0.1585826797 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 8 |
Hb_001755_040 |
0.1593998348 |
- |
- |
PREDICTED: cyclin-A2-4-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000352_010 |
0.1596253002 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 10 |
Hb_001862_050 |
0.1661380086 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 11 |
Hb_024973_020 |
0.1725405182 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 12 |
Hb_047659_010 |
0.1735594657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001141_050 |
0.1762687367 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002232_310 |
0.1767919275 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 15 |
Hb_000964_050 |
0.1792125253 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 16 |
Hb_011033_010 |
0.1794209945 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 17 |
Hb_002078_120 |
0.1794447039 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 18 |
Hb_000932_110 |
0.1799030494 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 19 |
Hb_071674_010 |
0.1812952895 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 20 |
Hb_003109_010 |
0.1851629665 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |