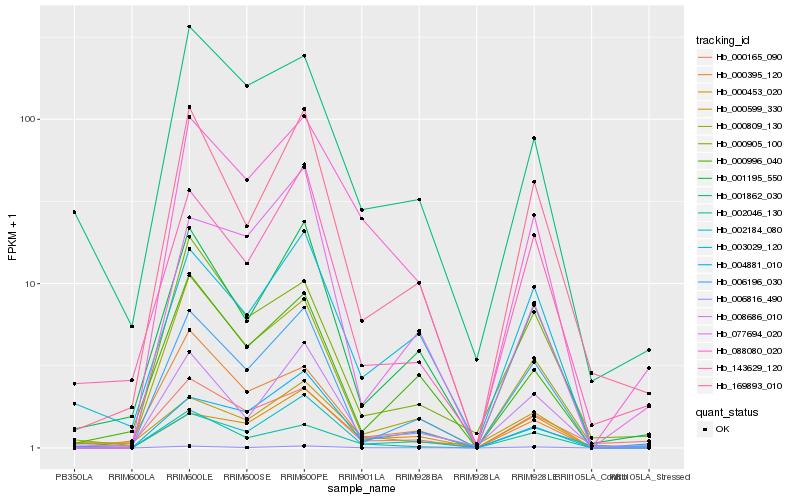
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_088080_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_004881_010 |
0.1612830946 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL1 [Jatropha curcas] |
| 3 |
Hb_006816_490 |
0.1635729037 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 21 [Jatropha curcas] |
| 4 |
Hb_001862_030 |
0.1845778944 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 5 |
Hb_002046_130 |
0.1850048333 |
- |
- |
PREDICTED: UDP-sugar transporter sqv-7-like [Jatropha curcas] |
| 6 |
Hb_169893_010 |
0.186543638 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 7 |
Hb_001195_550 |
0.1879717116 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000809_130 |
0.1927403324 |
- |
- |
PREDICTED: peroxidase 10 isoform X3 [Jatropha curcas] |
| 9 |
Hb_002184_080 |
0.1931518408 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 10 |
Hb_006196_030 |
0.1987531477 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 11 |
Hb_008686_010 |
0.199306756 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003029_120 |
0.2008291695 |
- |
- |
PREDICTED: uncharacterized protein LOC105642638 [Jatropha curcas] |
| 13 |
Hb_000996_040 |
0.202482811 |
- |
- |
hypothetical protein POPTR_0003s12500g [Populus trichocarpa] |
| 14 |
Hb_000599_330 |
0.2029063633 |
- |
- |
hypothetical protein JCGZ_17661 [Jatropha curcas] |
| 15 |
Hb_000395_120 |
0.2030794481 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 16 |
Hb_000453_020 |
0.2031250823 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000165_090 |
0.2041671566 |
- |
- |
PREDICTED: protein MID1-COMPLEMENTING ACTIVITY 2-like [Jatropha curcas] |
| 18 |
Hb_077694_020 |
0.2056831835 |
- |
- |
SAUR family protein [Theobroma cacao] |
| 19 |
Hb_143629_120 |
0.207136563 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 20 |
Hb_000905_100 |
0.2094315843 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |