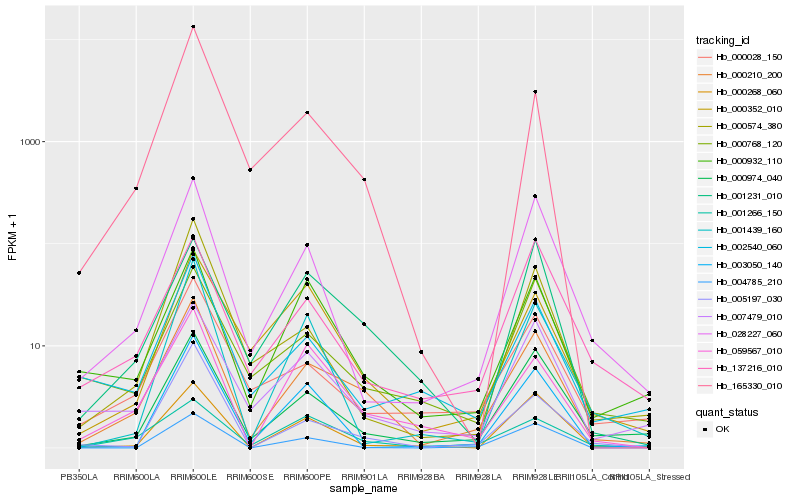
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000210_200 |
0.0 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 2 |
Hb_000974_040 |
0.1420296147 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Jatropha curcas] |
| 3 |
Hb_007479_010 |
0.1685738407 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 4 |
Hb_028227_060 |
0.1703519796 |
- |
- |
PREDICTED: non-specific lipid transfer protein GPI-anchored 1-like [Populus euphratica] |
| 5 |
Hb_000028_150 |
0.1709388358 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 6 |
Hb_165330_010 |
0.1774095632 |
- |
- |
BnaUnng00840D [Brassica napus] |
| 7 |
Hb_059567_010 |
0.179894265 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |
| 8 |
Hb_000932_110 |
0.182353547 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 9 |
Hb_001266_150 |
0.1827531516 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 10 |
Hb_000268_060 |
0.1843454741 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 11 |
Hb_005197_030 |
0.187403439 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH71-like isoform X2 [Populus euphratica] |
| 12 |
Hb_002540_060 |
0.188570588 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001231_010 |
0.1908478944 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 14 |
Hb_000574_380 |
0.1932329297 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 15 |
Hb_003050_140 |
0.1948025778 |
- |
- |
PREDICTED: potassium channel KAT1 [Jatropha curcas] |
| 16 |
Hb_000768_120 |
0.1949731844 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigB [Jatropha curcas] |
| 17 |
Hb_137216_010 |
0.1959328614 |
- |
- |
PREDICTED: unknown protein DS12 from 2D-PAGE of leaf, chloroplastic-like isoform X2 [Citrus sinensis] |
| 18 |
Hb_000352_010 |
0.1980505208 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 19 |
Hb_001439_160 |
0.199148851 |
- |
- |
PREDICTED: uncharacterized protein LOC105644097 [Jatropha curcas] |
| 20 |
Hb_004785_210 |
0.2026468228 |
- |
- |
hypothetical protein JCGZ_25215 [Jatropha curcas] |