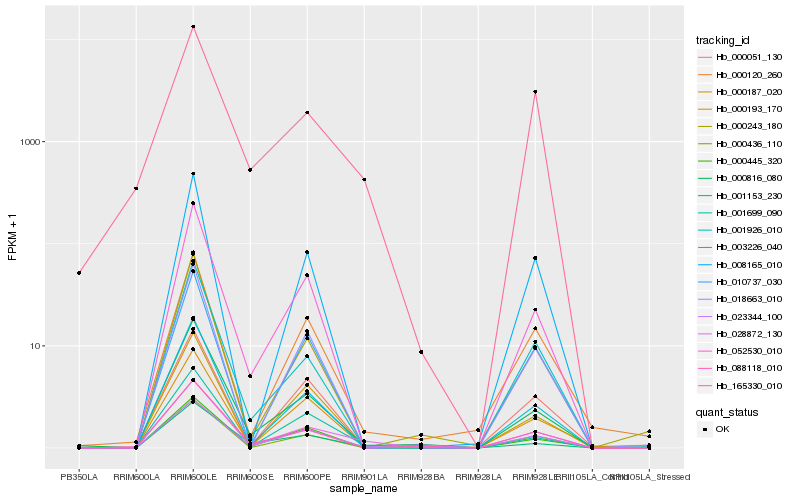
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028872_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644764 isoform X1 [Jatropha curcas] |
| 2 |
Hb_165330_010 |
0.1300150979 |
- |
- |
BnaUnng00840D [Brassica napus] |
| 3 |
Hb_001153_230 |
0.1419419358 |
- |
- |
PREDICTED: uncharacterized protein LOC105645885 [Jatropha curcas] |
| 4 |
Hb_000816_080 |
0.142830055 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 5 |
Hb_052530_010 |
0.1461079886 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000436_110 |
0.1488946742 |
- |
- |
PREDICTED: uncharacterized protein LOC105141197 [Populus euphratica] |
| 7 |
Hb_003226_040 |
0.150808379 |
- |
- |
PREDICTED: endoglucanase 17 [Populus euphratica] |
| 8 |
Hb_000120_260 |
0.1510251326 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 9 |
Hb_000445_320 |
0.1555025059 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like isoform X2 [Gossypium raimondii] |
| 10 |
Hb_001926_010 |
0.1559836876 |
- |
- |
hypothetical protein Csa_2G383910 [Cucumis sativus] |
| 11 |
Hb_001699_090 |
0.1572623665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_088118_010 |
0.1584378706 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 13 |
Hb_008165_010 |
0.1591584498 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein [Solanum lycopersicum] |
| 14 |
Hb_010737_030 |
0.1618190794 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000193_170 |
0.1622001829 |
transcription factor |
TF Family: MYB |
Myb domain 16-like protein [Theobroma cacao] |
| 16 |
Hb_018663_010 |
0.1623196015 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |
| 17 |
Hb_000187_020 |
0.1625210576 |
- |
- |
Xanthine/uracil permease family protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_000243_180 |
0.1636680229 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1 [Jatropha curcas] |
| 19 |
Hb_000051_130 |
0.1647703507 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 20 |
Hb_023344_100 |
0.1657431746 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.2 [Jatropha curcas] |