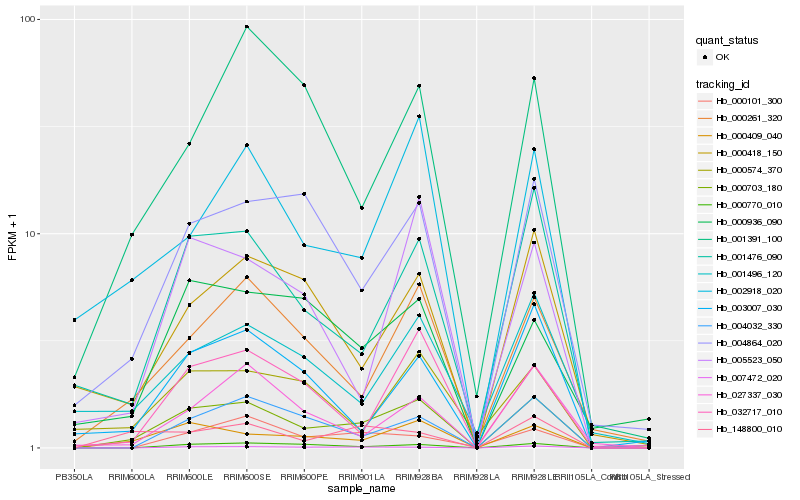
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000703_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 2 |
Hb_000409_040 |
0.139835569 |
transcription factor |
TF Family: AP2 |
hypothetical protein POPTR_0017s11470g [Populus trichocarpa] |
| 3 |
Hb_000261_320 |
0.1767229325 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_001476_090 |
0.1797804636 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 5 |
Hb_004864_020 |
0.1841431432 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 6 |
Hb_002918_020 |
0.1884965674 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007472_020 |
0.1890210768 |
- |
- |
hypothetical protein PRUPE_ppa016115mg [Prunus persica] |
| 8 |
Hb_000770_010 |
0.1908191573 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 9 |
Hb_004032_330 |
0.2005370529 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_032717_010 |
0.2071610987 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_001496_120 |
0.2120982315 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_005523_050 |
0.2127105045 |
- |
- |
hypothetical protein JCGZ_09175 [Jatropha curcas] |
| 13 |
Hb_000418_150 |
0.212872953 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_148800_010 |
0.2133874313 |
- |
- |
- |
| 15 |
Hb_003007_030 |
0.217072178 |
- |
- |
- |
| 16 |
Hb_001391_100 |
0.2195994304 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 17 |
Hb_000936_090 |
0.2227797538 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X2 [Jatropha curcas] |
| 18 |
Hb_000101_300 |
0.2243458555 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein B456_002G046500, partial [Gossypium raimondii] |
| 19 |
Hb_000574_370 |
0.2244668335 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 20 |
Hb_027337_030 |
0.2245953785 |
- |
- |
PREDICTED: uncharacterized protein LOC104602841 [Nelumbo nucifera] |