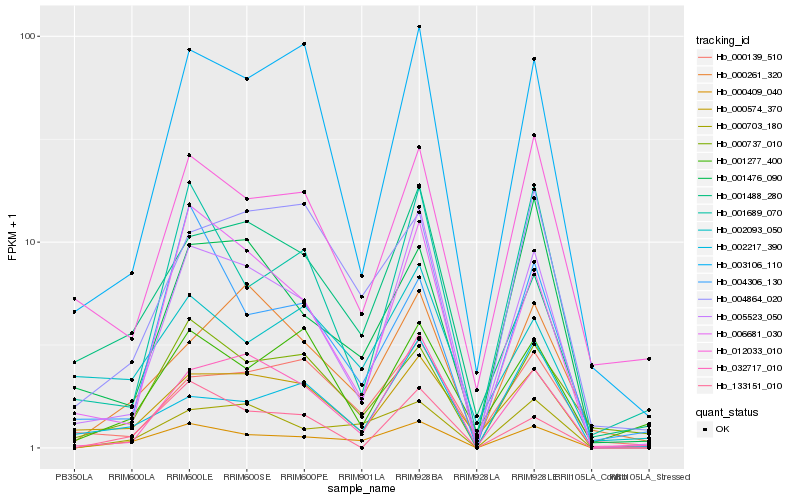
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000409_040 |
0.0 |
transcription factor |
TF Family: AP2 |
hypothetical protein POPTR_0017s11470g [Populus trichocarpa] |
| 2 |
Hb_000703_180 |
0.139835569 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 3 |
Hb_005523_050 |
0.1801391862 |
- |
- |
hypothetical protein JCGZ_09175 [Jatropha curcas] |
| 4 |
Hb_000737_010 |
0.1986172268 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X1 [Jatropha curcas] |
| 5 |
Hb_000574_370 |
0.204941285 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 6 |
Hb_004864_020 |
0.2074135428 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 7 |
Hb_006681_030 |
0.2109462438 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Populus euphratica] |
| 8 |
Hb_032717_010 |
0.2140491306 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_001689_070 |
0.2148274161 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_003106_110 |
0.2149330874 |
- |
- |
betaine aldehyde dehydrogenase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_133151_010 |
0.2168109129 |
- |
- |
NBS-LRR resistance gene-like protein ARGH30 [Populus trichocarpa] |
| 12 |
Hb_001488_280 |
0.2202822067 |
- |
- |
hypothetical protein JCGZ_19947 [Jatropha curcas] |
| 13 |
Hb_002093_050 |
0.2209072381 |
- |
- |
hypothetical protein POPTR_0008s02940g [Populus trichocarpa] |
| 14 |
Hb_012033_010 |
0.2215951756 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 15 |
Hb_000261_320 |
0.2225209939 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_001476_090 |
0.2228449513 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 17 |
Hb_004306_130 |
0.2234776584 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002217_390 |
0.2247510295 |
- |
- |
PREDICTED: galactokinase [Jatropha curcas] |
| 19 |
Hb_000139_510 |
0.226329009 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001277_400 |
0.2280961978 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 4B isoform X3 [Jatropha curcas] |