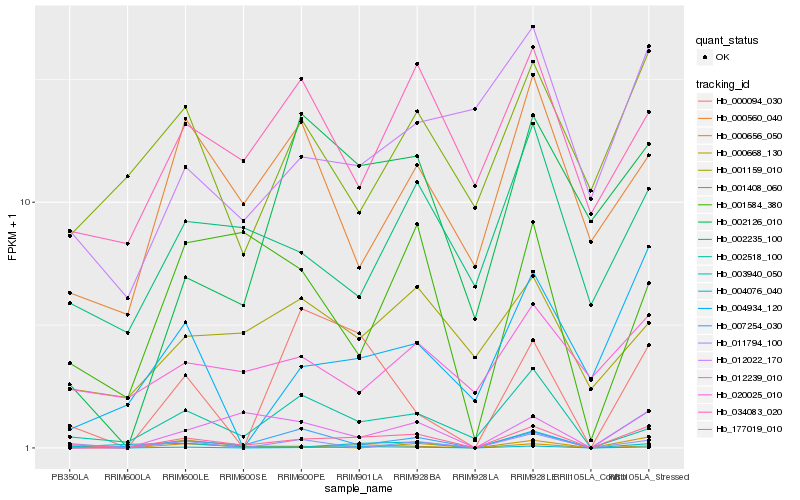
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_177019_010 |
0.0 |
- |
- |
Uncharacterized protein TCM_004012 [Theobroma cacao] |
| 2 |
Hb_004076_040 |
0.2553481194 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_000656_050 |
0.2750973388 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase II, chloroplastic-like [Vitis vinifera] |
| 4 |
Hb_004934_120 |
0.2916404667 |
- |
- |
- |
| 5 |
Hb_007254_030 |
0.2921619267 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_000668_130 |
0.2922440685 |
- |
- |
- |
| 7 |
Hb_002518_100 |
0.2930757636 |
- |
- |
- |
| 8 |
Hb_012239_010 |
0.2942744997 |
- |
- |
- |
| 9 |
Hb_002126_010 |
0.2986015531 |
- |
- |
predicted protein [Hordeum vulgare subsp. vulgare] |
| 10 |
Hb_000094_030 |
0.3021844912 |
- |
- |
- |
| 11 |
Hb_002235_100 |
0.304064701 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 4 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001408_060 |
0.3105385923 |
- |
- |
PREDICTED: 60S ribosomal protein L3 [Eucalyptus grandis] |
| 13 |
Hb_001584_380 |
0.3122170665 |
- |
- |
PREDICTED: tropomyosin-like [Jatropha curcas] |
| 14 |
Hb_003940_050 |
0.3139859652 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 15 |
Hb_034083_020 |
0.3156782579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_011794_100 |
0.3168216838 |
- |
- |
PREDICTED: seven transmembrane domain-containing tyrosine-protein kinase 1-like [Jatropha curcas] |
| 17 |
Hb_012022_170 |
0.3183514222 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 18 |
Hb_020025_010 |
0.3204550841 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 19 |
Hb_000560_040 |
0.3212776767 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001159_010 |
0.3214845847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |