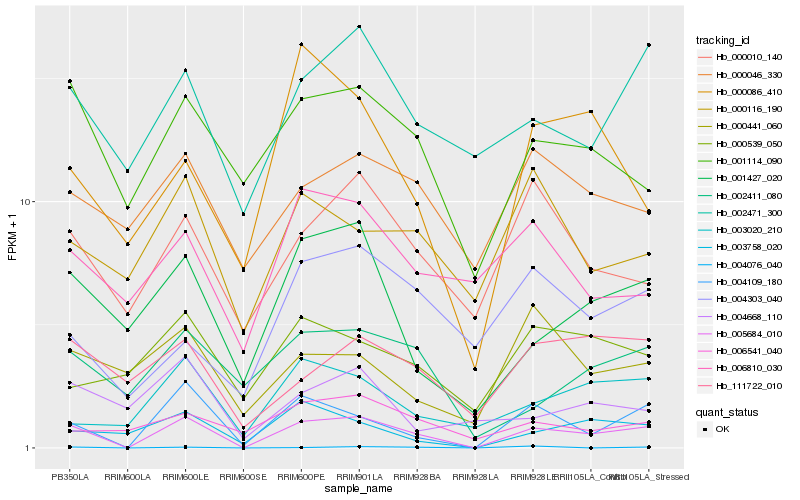
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005684_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104216799 [Nicotiana sylvestris] |
| 2 |
Hb_004109_180 |
0.1910272374 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001427_020 |
0.2540751293 |
- |
- |
hypothetical protein JCGZ_07310 [Jatropha curcas] |
| 4 |
Hb_003020_210 |
0.2664683073 |
- |
- |
Structural maintenance of chromosomes 2-2 -like protein [Gossypium arboreum] |
| 5 |
Hb_003758_020 |
0.2691064346 |
- |
- |
UGTPg34 [Panax ginseng] |
| 6 |
Hb_111722_010 |
0.2723419014 |
- |
- |
PREDICTED: uncharacterized protein LOC105634044 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002471_300 |
0.2730782774 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein Sm D2-like [Jatropha curcas] |
| 8 |
Hb_000010_140 |
0.2747770887 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 11, chloroplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_001114_090 |
0.2751964024 |
- |
- |
PREDICTED: magnesium-dependent phosphatase 1 [Jatropha curcas] |
| 10 |
Hb_002411_080 |
0.2767186623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000086_410 |
0.2832264928 |
- |
- |
hypothetical protein CISIN_1g0263261mg [Citrus sinensis] |
| 12 |
Hb_004303_040 |
0.2877447022 |
- |
- |
PREDICTED: fumarate hydratase 1, mitochondrial [Jatropha curcas] |
| 13 |
Hb_006541_040 |
0.2915378562 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 2 [Jatropha curcas] |
| 14 |
Hb_000116_190 |
0.2934838696 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000441_060 |
0.2953383041 |
- |
- |
PREDICTED: glycolipid transfer protein [Jatropha curcas] |
| 16 |
Hb_000539_050 |
0.2961455857 |
- |
- |
bile acid:sodium symporter, putative [Ricinus communis] |
| 17 |
Hb_000046_330 |
0.2972061437 |
- |
- |
PREDICTED: uncharacterized protein LOC105631826 [Jatropha curcas] |
| 18 |
Hb_006810_030 |
0.2974936716 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_004076_040 |
0.2983964339 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_004668_110 |
0.2994377217 |
- |
- |
conserved hypothetical protein [Ricinus communis] |