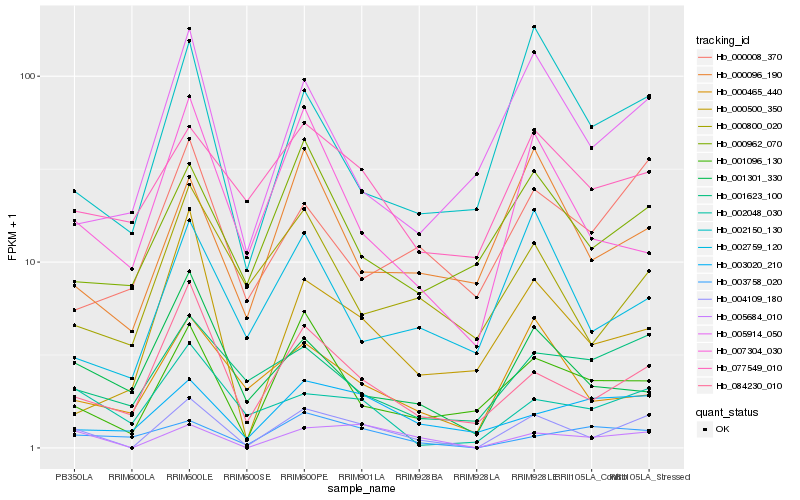
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004109_180 |
0.0 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005684_010 |
0.1910272374 |
- |
- |
PREDICTED: uncharacterized protein LOC104216799 [Nicotiana sylvestris] |
| 3 |
Hb_084230_010 |
0.2220921942 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 4 |
Hb_001096_130 |
0.2423125141 |
- |
- |
PREDICTED: RING-H2 finger protein ATL7-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_007304_030 |
0.257545297 |
- |
- |
PREDICTED: uncharacterized protein LOC105635926 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002150_130 |
0.2624084271 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_002048_030 |
0.2632975529 |
- |
- |
PREDICTED: putative quinone-oxidoreductase homolog, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000465_440 |
0.2641146707 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2-like [Jatropha curcas] |
| 9 |
Hb_005914_050 |
0.2658906136 |
- |
- |
PREDICTED: 50S ribosomal protein L28, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000096_190 |
0.2661071311 |
- |
- |
unknown [Lotus japonicus] |
| 11 |
Hb_003020_210 |
0.2691814099 |
- |
- |
Structural maintenance of chromosomes 2-2 -like protein [Gossypium arboreum] |
| 12 |
Hb_000962_070 |
0.2704886357 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 13, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_000500_350 |
0.2717039328 |
- |
- |
PREDICTED: uncharacterized protein LOC105649734 [Jatropha curcas] |
| 14 |
Hb_001623_100 |
0.2725709657 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 15 |
Hb_003758_020 |
0.2739239322 |
- |
- |
UGTPg34 [Panax ginseng] |
| 16 |
Hb_000800_020 |
0.2741010643 |
- |
- |
PREDICTED: 3-phosphoinositide-dependent protein kinase 2 [Jatropha curcas] |
| 17 |
Hb_001301_330 |
0.2750085787 |
- |
- |
OTU-like cysteine protease family protein [Populus trichocarpa] |
| 18 |
Hb_002759_120 |
0.2750689108 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_077549_010 |
0.2757820598 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000008_370 |
0.2778838515 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |