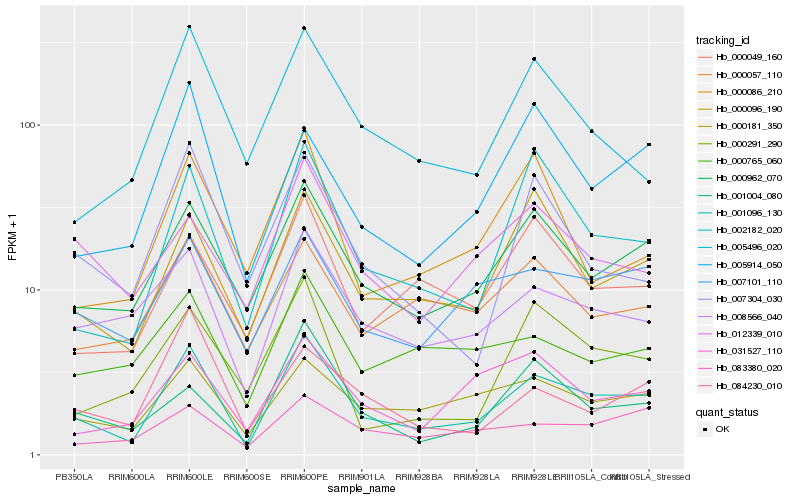
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001096_130 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL7-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000962_070 |
0.1440750087 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 13, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_002182_020 |
0.1528042259 |
- |
- |
Thylakoid lumenal 17.4 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_008566_040 |
0.1552152409 |
- |
- |
PREDICTED: uncharacterized protein LOC105647590 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000096_190 |
0.1592801309 |
- |
- |
unknown [Lotus japonicus] |
| 6 |
Hb_000181_350 |
0.1636594041 |
- |
- |
PREDICTED: uncharacterized protein LOC105111090 [Populus euphratica] |
| 7 |
Hb_007101_110 |
0.169022088 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 7 [Jatropha curcas] |
| 8 |
Hb_000049_160 |
0.1794615625 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |
| 9 |
Hb_001004_080 |
0.1801034562 |
- |
- |
hypothetical protein POPTR_0014s14130g [Populus trichocarpa] |
| 10 |
Hb_005496_020 |
0.1802425651 |
- |
- |
PREDICTED: uncharacterized protein LOC105630867 [Jatropha curcas] |
| 11 |
Hb_000765_060 |
0.1805602037 |
- |
- |
Alcohol dehydrogenase-like 6 [Glycine soja] |
| 12 |
Hb_031527_110 |
0.1836932525 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Jatropha curcas] |
| 13 |
Hb_005914_050 |
0.1844352357 |
- |
- |
PREDICTED: 50S ribosomal protein L28, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000057_110 |
0.1854104077 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 15 |
Hb_007304_030 |
0.1856858923 |
- |
- |
PREDICTED: uncharacterized protein LOC105635926 isoform X2 [Jatropha curcas] |
| 16 |
Hb_083380_020 |
0.1866701461 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 17 |
Hb_000086_210 |
0.1883540282 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_000291_290 |
0.1904137414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_012339_010 |
0.1904288892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_084230_010 |
0.1910749762 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |