| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
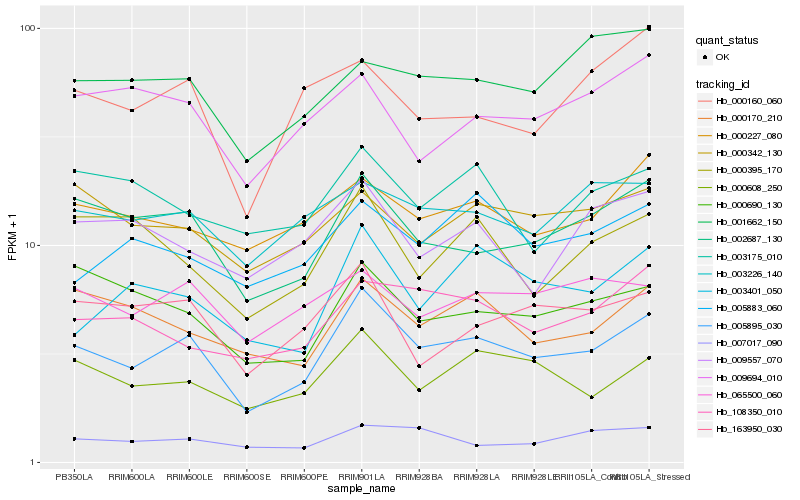
Hb_005895_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643388 [Jatropha curcas] |
| 2 |
Hb_002687_130 |
0.1009044936 |
- |
- |
PREDICTED: uncharacterized protein LOC105631153 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000608_250 |
0.1109082902 |
- |
- |
Cytidine/deoxycytidylate deaminase family protein isoform 2 [Theobroma cacao] |
| 4 |
Hb_000170_210 |
0.1244072611 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_003401_050 |
0.1246718373 |
- |
- |
PREDICTED: origin of replication complex subunit 2 [Jatropha curcas] |
| 6 |
Hb_163950_030 |
0.124924066 |
- |
- |
PREDICTED: uncharacterized protein LOC105648509 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000690_130 |
0.1258622327 |
- |
- |
PREDICTED: uncharacterized protein LOC105631771 [Jatropha curcas] |
| 8 |
Hb_000227_080 |
0.12639492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_108350_010 |
0.1267813286 |
- |
- |
GTP-binding protein erg, putative [Ricinus communis] |
| 10 |
Hb_003226_140 |
0.1273219367 |
- |
- |
small nuclear ribonucleoprotein U)1a,U)2b, putative [Ricinus communis] |
| 11 |
Hb_065500_060 |
0.1283814082 |
- |
- |
PREDICTED: uncharacterized protein LOC105633723 [Jatropha curcas] |
| 12 |
Hb_000342_130 |
0.1308077029 |
- |
- |
PREDICTED: uncharacterized protein LOC105638175 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007017_090 |
0.1317394412 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 186 [Jatropha curcas] |
| 14 |
Hb_000395_170 |
0.1326768835 |
- |
- |
PREDICTED: probable transcriptional regulatory protein At2g25830 [Jatropha curcas] |
| 15 |
Hb_001662_150 |
0.1352181129 |
- |
- |
Scaffold attachment factor B1 [Medicago truncatula] |
| 16 |
Hb_003175_010 |
0.1357829506 |
- |
- |
PREDICTED: 39S ribosomal protein L4, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000160_060 |
0.1361829173 |
- |
- |
unknown [Populus trichocarpa] |
| 18 |
Hb_005883_060 |
0.136866239 |
- |
- |
PREDICTED: polynucleotide 5'-hydroxyl-kinase NOL9 isoform X1 [Jatropha curcas] |
| 19 |
Hb_009557_070 |
0.1369236887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_009694_010 |
0.1373229566 |
- |
- |
conserved hypothetical protein [Ricinus communis] |