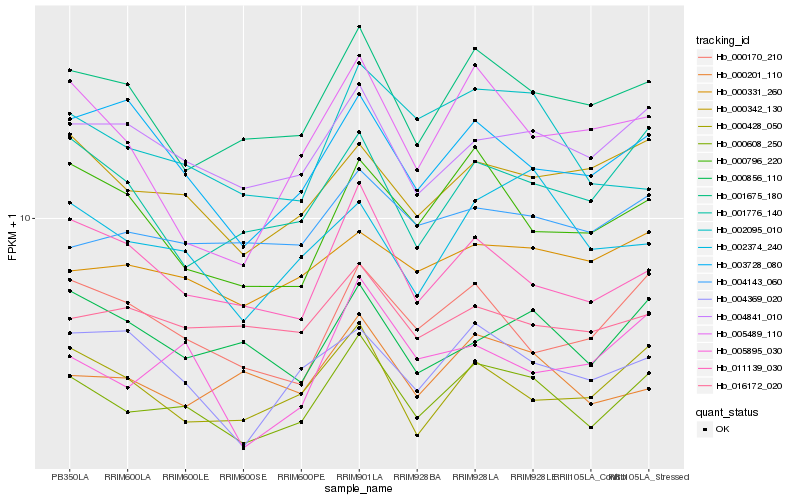
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000608_250 |
0.0 |
- |
- |
Cytidine/deoxycytidylate deaminase family protein isoform 2 [Theobroma cacao] |
| 2 |
Hb_001675_180 |
0.0918488305 |
- |
- |
PREDICTED: golgin candidate 5 [Jatropha curcas] |
| 3 |
Hb_004841_010 |
0.0940899366 |
- |
- |
PREDICTED: 60S ribosomal protein L2, mitochondrial [Jatropha curcas] |
| 4 |
Hb_011139_030 |
0.097096433 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR2-like [Jatropha curcas] |
| 5 |
Hb_000342_130 |
0.102423537 |
- |
- |
PREDICTED: uncharacterized protein LOC105638175 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000428_050 |
0.1030905659 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
PREDICTED: tRNA dimethylallyltransferase 9 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000170_210 |
0.1066223033 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000796_220 |
0.1073360584 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X3 [Jatropha curcas] |
| 9 |
Hb_001776_140 |
0.1073780148 |
- |
- |
PREDICTED: probable GTP-binding protein OBGM, mitochondrial isoform X2 [Jatropha curcas] |
| 10 |
Hb_002095_010 |
0.1088025899 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002374_240 |
0.1089442266 |
- |
- |
nucellin, putative [Ricinus communis] |
| 12 |
Hb_000201_110 |
0.1099828375 |
- |
- |
hypothetical protein JCGZ_21690 [Jatropha curcas] |
| 13 |
Hb_003728_080 |
0.1102013529 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3-like [Jatropha curcas] |
| 14 |
Hb_005895_030 |
0.1109082902 |
- |
- |
PREDICTED: uncharacterized protein LOC105643388 [Jatropha curcas] |
| 15 |
Hb_000331_260 |
0.1121192283 |
- |
- |
PREDICTED: WD repeat-containing protein 74 isoform X2 [Jatropha curcas] |
| 16 |
Hb_004143_060 |
0.1125947957 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_000856_110 |
0.1126248331 |
- |
- |
PREDICTED: uncharacterized protein LOC105640477 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004369_020 |
0.1137165484 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 19 |
Hb_005489_110 |
0.1148695198 |
- |
- |
PREDICTED: putative transferase CAF17 homolog, mitochondrial [Jatropha curcas] |
| 20 |
Hb_016172_020 |
0.115372156 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |