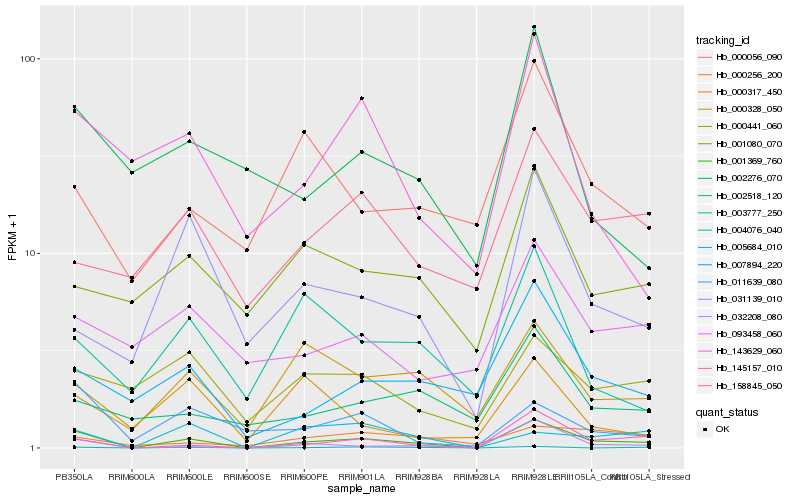
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_760 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa019417mg [Prunus persica] |
| 2 |
Hb_145157_010 |
0.2266751987 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 3 |
Hb_032208_080 |
0.2478320228 |
- |
- |
PREDICTED: phytoene synthase 2, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_004076_040 |
0.2678951758 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_007894_220 |
0.2741846473 |
- |
- |
PREDICTED: probable folate-biopterin transporter 4 [Jatropha curcas] |
| 6 |
Hb_011639_080 |
0.2901786227 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 7 |
Hb_143629_060 |
0.2927244799 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_031139_010 |
0.299250335 |
- |
- |
PREDICTED: uncharacterized protein LOC105632403 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005684_010 |
0.3026724078 |
- |
- |
PREDICTED: uncharacterized protein LOC104216799 [Nicotiana sylvestris] |
| 10 |
Hb_000441_060 |
0.3050384661 |
- |
- |
PREDICTED: glycolipid transfer protein [Jatropha curcas] |
| 11 |
Hb_002518_120 |
0.305384138 |
- |
- |
hypothetical protein RCOM_0731250 [Ricinus communis] |
| 12 |
Hb_002276_070 |
0.306373317 |
- |
- |
PREDICTED: heat shock protein 83 [Jatropha curcas] |
| 13 |
Hb_158845_050 |
0.3071340389 |
- |
- |
PREDICTED: uncharacterized protein LOC105122890 [Populus euphratica] |
| 14 |
Hb_000328_050 |
0.3073499858 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_093458_060 |
0.3074197645 |
- |
- |
PREDICTED: uncharacterized protein LOC105633194 [Jatropha curcas] |
| 16 |
Hb_003777_250 |
0.3081402438 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 17 |
Hb_001080_070 |
0.3090435483 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 18 |
Hb_000056_090 |
0.3091311889 |
- |
- |
UDP-galactose transporter 3 isoform 1 [Theobroma cacao] |
| 19 |
Hb_000256_200 |
0.3113094085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000317_450 |
0.3125302947 |
- |
- |
PREDICTED: uncharacterized protein LOC105647032 isoform X1 [Jatropha curcas] |