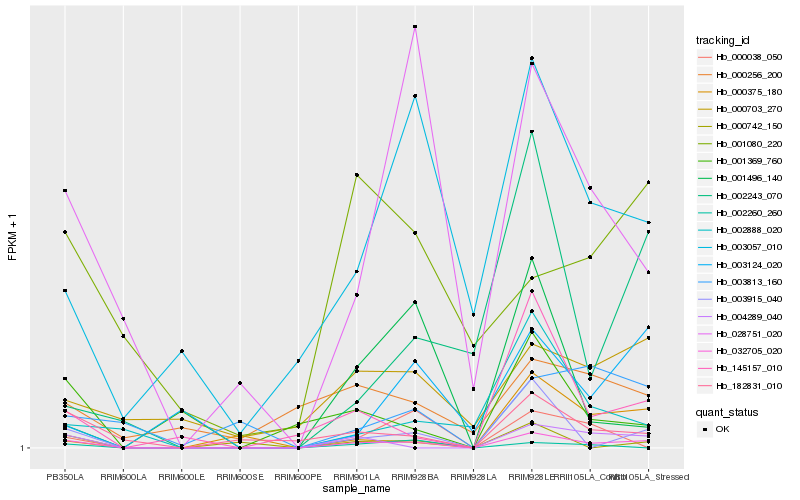
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032705_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 2 |
Hb_003915_040 |
0.2727435744 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At2g30600 isoform X1 [Jatropha curcas] |
| 3 |
Hb_145157_010 |
0.2756401853 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 4 |
Hb_004289_040 |
0.2761936545 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 5 |
Hb_182831_010 |
0.2891629221 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 6 |
Hb_028751_020 |
0.3077162166 |
- |
- |
PREDICTED: SCY1-like protein 2 [Jatropha curcas] |
| 7 |
Hb_001496_140 |
0.3175398572 |
- |
- |
- |
| 8 |
Hb_000742_150 |
0.3221495728 |
- |
- |
- |
| 9 |
Hb_003057_010 |
0.3265035354 |
- |
- |
hypothetical protein JCGZ_06666 [Jatropha curcas] |
| 10 |
Hb_001369_760 |
0.3362199453 |
- |
- |
hypothetical protein PRUPE_ppa019417mg [Prunus persica] |
| 11 |
Hb_002243_070 |
0.3383963059 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 12 |
Hb_002888_020 |
0.3404797937 |
- |
- |
- |
| 13 |
Hb_000375_180 |
0.3408194249 |
- |
- |
PREDICTED: uncharacterized protein LOC104593656 [Nelumbo nucifera] |
| 14 |
Hb_000703_270 |
0.3421430859 |
- |
- |
hypothetical protein B456_012G039600 [Gossypium raimondii] |
| 15 |
Hb_003124_020 |
0.3485265705 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105111046 [Populus euphratica] |
| 16 |
Hb_002260_260 |
0.3533904992 |
- |
- |
hypothetical protein VITISV_037667 [Vitis vinifera] |
| 17 |
Hb_003813_160 |
0.3562337656 |
- |
- |
- |
| 18 |
Hb_000256_200 |
0.3568755535 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001080_220 |
0.3619409639 |
- |
- |
TPA: hypothetical protein ZEAMMB73_966926 [Zea mays] |
| 20 |
Hb_000038_050 |
0.3672908736 |
- |
- |
- |