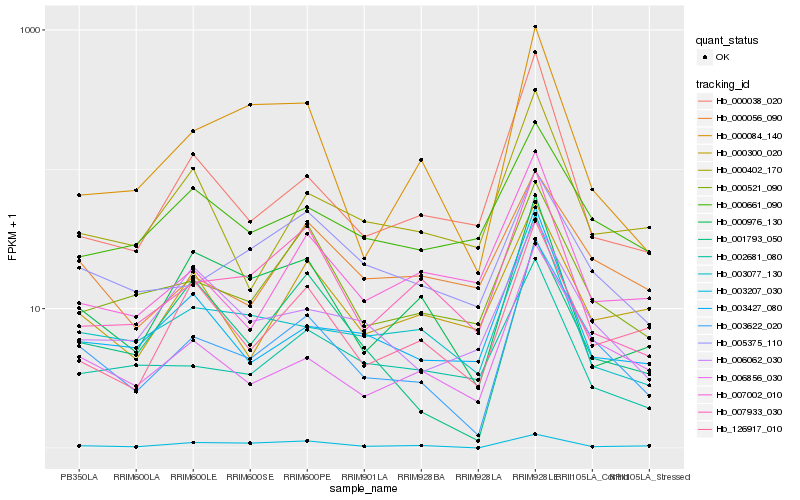
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003622_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At5g39865-like [Jatropha curcas] |
| 2 |
Hb_006856_030 |
0.1427243486 |
- |
- |
Uridine kinase-like protein 4 [Glycine soja] |
| 3 |
Hb_000521_090 |
0.1686377556 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 4 |
Hb_126917_010 |
0.1778195728 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 5 |
Hb_003077_130 |
0.18179645 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |
| 6 |
Hb_003427_080 |
0.1823093649 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002681_080 |
0.1850656453 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007933_030 |
0.1861211731 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 9 |
Hb_000056_090 |
0.1878502208 |
- |
- |
UDP-galactose transporter 3 isoform 1 [Theobroma cacao] |
| 10 |
Hb_000084_140 |
0.1884481641 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 11 |
Hb_005375_110 |
0.1889695423 |
- |
- |
metal tolerance protein 1 [Populus trichocarpa x Populus deltoides] |
| 12 |
Hb_000300_020 |
0.1901289237 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000402_170 |
0.1915600286 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 14 |
Hb_001793_050 |
0.193325511 |
- |
- |
PREDICTED: uncharacterized protein LOC105632663 [Jatropha curcas] |
| 15 |
Hb_000976_130 |
0.1952197355 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 16 |
Hb_003207_030 |
0.1953572943 |
- |
- |
hypothetical protein VITISV_027754 [Vitis vinifera] |
| 17 |
Hb_000038_020 |
0.1957425243 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_006062_030 |
0.1957446516 |
- |
- |
PREDICTED: translation initiation factor IF-2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000661_090 |
0.1971425925 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 20 |
Hb_007002_010 |
0.1983389777 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |