| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
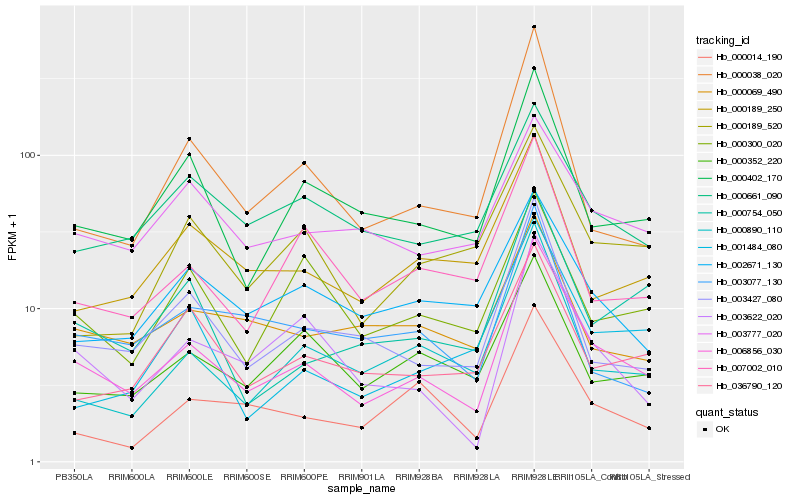
Hb_006856_030 |
0.0 |
- |
- |
Uridine kinase-like protein 4 [Glycine soja] |
| 2 |
Hb_000402_170 |
0.1384472045 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 3 |
Hb_003427_080 |
0.1389724637 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003622_020 |
0.1427243486 |
- |
- |
PREDICTED: uncharacterized protein At5g39865-like [Jatropha curcas] |
| 5 |
Hb_000890_110 |
0.155209399 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |
| 6 |
Hb_000754_050 |
0.1584715393 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003777_020 |
0.1617900158 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 8 |
Hb_036790_120 |
0.1637652042 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000038_020 |
0.1639694684 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_007002_010 |
0.1646691665 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 11 |
Hb_000661_090 |
0.1653284394 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000352_220 |
0.1666586676 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000014_190 |
0.1669259288 |
- |
- |
Phytoene dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_000189_250 |
0.1672267815 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002671_130 |
0.1689749804 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 16 |
Hb_000300_020 |
0.1701507673 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000069_490 |
0.1705296998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000189_520 |
0.1720891886 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 19 |
Hb_001484_080 |
0.1727172922 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 20 |
Hb_003077_130 |
0.1730883998 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |