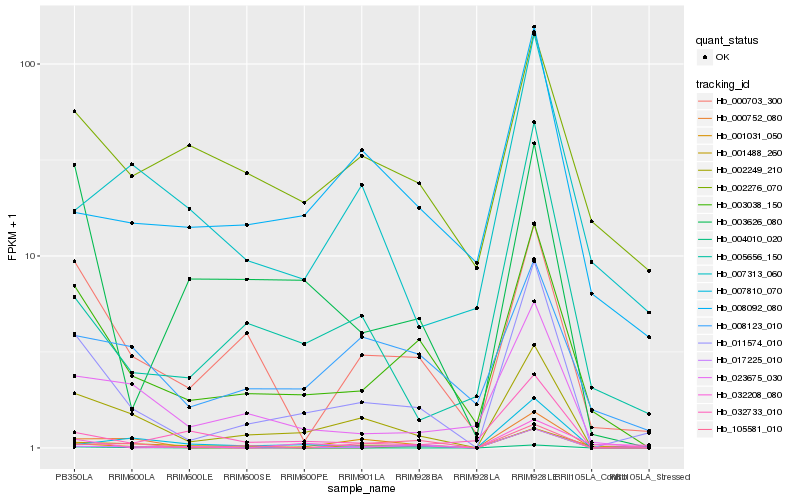
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011574_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_002249_210 |
0.1604920473 |
- |
- |
- |
| 3 |
Hb_003038_150 |
0.1996031589 |
- |
- |
- |
| 4 |
Hb_000703_300 |
0.2403420382 |
- |
- |
- |
| 5 |
Hb_005656_150 |
0.2492131844 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 6 |
Hb_023675_030 |
0.2578466346 |
- |
- |
- |
| 7 |
Hb_001488_260 |
0.2677516721 |
- |
- |
Alpha-2-macroglobulin [Gossypium arboreum] |
| 8 |
Hb_003626_080 |
0.2755340283 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_032733_010 |
0.2853924513 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 10 |
Hb_001031_050 |
0.2861242299 |
- |
- |
- |
| 11 |
Hb_032208_080 |
0.2873892929 |
- |
- |
PREDICTED: phytoene synthase 2, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_007810_070 |
0.2886878778 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 13 |
Hb_008092_080 |
0.2899198751 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 14 |
Hb_008123_010 |
0.2934365769 |
- |
- |
- |
| 15 |
Hb_017225_010 |
0.2971564558 |
- |
- |
PREDICTED: nodulation receptor kinase-like [Jatropha curcas] |
| 16 |
Hb_004010_020 |
0.2977251039 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 17 |
Hb_105581_010 |
0.3033100921 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 18 |
Hb_002276_070 |
0.3092657452 |
- |
- |
PREDICTED: heat shock protein 83 [Jatropha curcas] |
| 19 |
Hb_007313_060 |
0.3093083947 |
- |
- |
PREDICTED: paramyosin [Jatropha curcas] |
| 20 |
Hb_000752_080 |
0.3094622068 |
- |
- |
- |