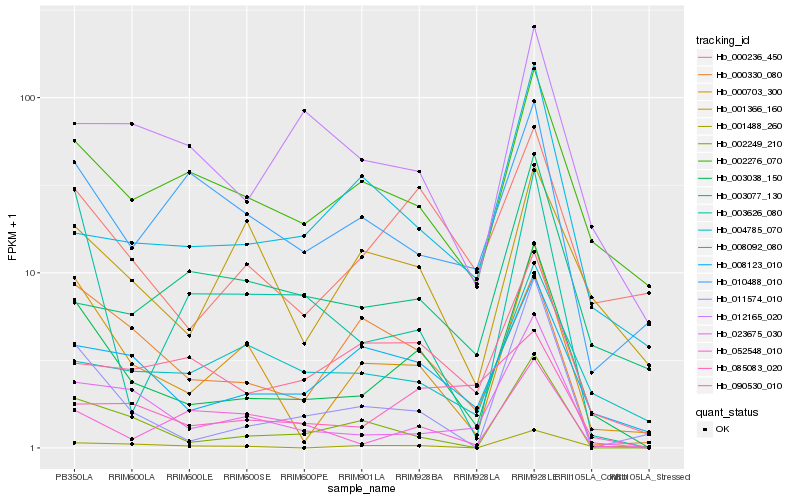
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003038_150 |
0.0 |
- |
- |
- |
| 2 |
Hb_000703_300 |
0.191349551 |
- |
- |
- |
| 3 |
Hb_011574_010 |
0.1996031589 |
- |
- |
- |
| 4 |
Hb_023675_030 |
0.210152682 |
- |
- |
- |
| 5 |
Hb_002249_210 |
0.2138577758 |
- |
- |
- |
| 6 |
Hb_000236_450 |
0.2209885238 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 7 |
Hb_008123_010 |
0.2216730079 |
- |
- |
- |
| 8 |
Hb_002276_070 |
0.229778023 |
- |
- |
PREDICTED: heat shock protein 83 [Jatropha curcas] |
| 9 |
Hb_003626_080 |
0.2409640713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001488_260 |
0.24471715 |
- |
- |
Alpha-2-macroglobulin [Gossypium arboreum] |
| 11 |
Hb_090530_010 |
0.2479565506 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 12 |
Hb_012165_020 |
0.2591649843 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |
| 13 |
Hb_000330_080 |
0.2618821102 |
- |
- |
auxilin, putative [Ricinus communis] |
| 14 |
Hb_008092_080 |
0.2621648064 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 15 |
Hb_052548_010 |
0.2648870186 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 16 |
Hb_004785_070 |
0.2677484434 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_001366_160 |
0.2715621816 |
- |
- |
- |
| 18 |
Hb_003077_130 |
0.2728143529 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |
| 19 |
Hb_010488_010 |
0.2729512038 |
- |
- |
PREDICTED: (+)-neomenthol dehydrogenase-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_085083_020 |
0.2773066391 |
- |
- |
PREDICTED: receptor-like protein kinase THESEUS 1 [Jatropha curcas] |