| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
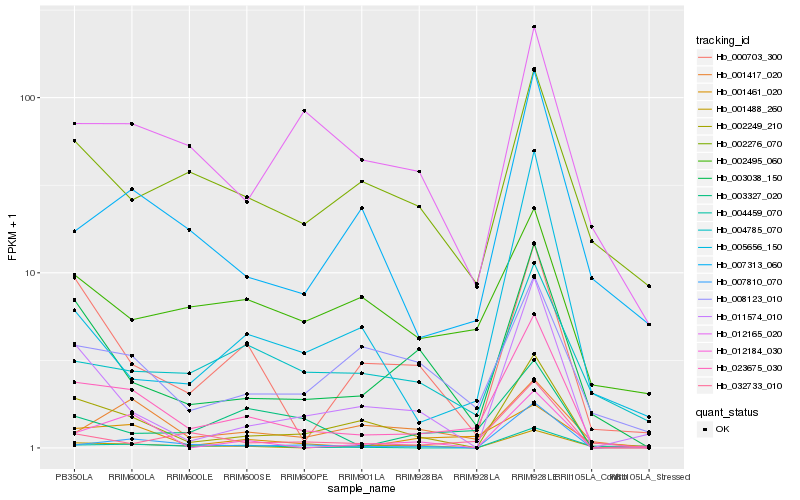
Hb_023675_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_003038_150 |
0.210152682 |
- |
- |
- |
| 3 |
Hb_001461_020 |
0.2108240935 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_002249_210 |
0.2130485946 |
- |
- |
- |
| 5 |
Hb_007313_060 |
0.2294718515 |
- |
- |
PREDICTED: paramyosin [Jatropha curcas] |
| 6 |
Hb_000703_300 |
0.2455788424 |
- |
- |
- |
| 7 |
Hb_032733_010 |
0.2486165545 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 8 |
Hb_003327_020 |
0.2565301807 |
- |
- |
PREDICTED: phytosulfokine receptor 2-like, partial [Jatropha curcas] |
| 9 |
Hb_008123_010 |
0.2575247771 |
- |
- |
- |
| 10 |
Hb_011574_010 |
0.2578466346 |
- |
- |
- |
| 11 |
Hb_001417_020 |
0.2583731518 |
- |
- |
PREDICTED: uncharacterized protein LOC104799692 [Tarenaya hassleriana] |
| 12 |
Hb_004785_070 |
0.2614554954 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_005656_150 |
0.2620766572 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 14 |
Hb_002276_070 |
0.2633581882 |
- |
- |
PREDICTED: heat shock protein 83 [Jatropha curcas] |
| 15 |
Hb_004459_070 |
0.2650424217 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 16 |
Hb_012165_020 |
0.2655275969 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |
| 17 |
Hb_001488_260 |
0.2658476768 |
- |
- |
Alpha-2-macroglobulin [Gossypium arboreum] |
| 18 |
Hb_002495_060 |
0.2702386889 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_012184_030 |
0.2703543844 |
- |
- |
- |
| 20 |
Hb_007810_070 |
0.2705933303 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |