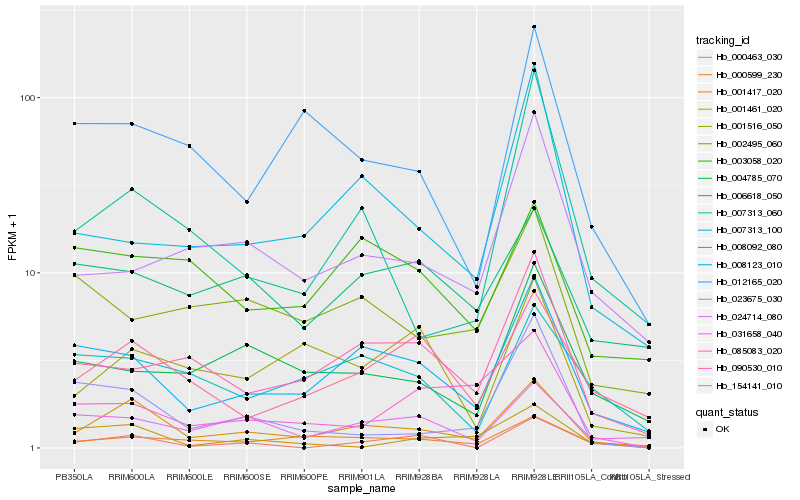
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001417_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104799692 [Tarenaya hassleriana] |
| 2 |
Hb_008123_010 |
0.1936711212 |
- |
- |
- |
| 3 |
Hb_154141_010 |
0.2194576773 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 4 |
Hb_000599_230 |
0.2207725018 |
- |
- |
- |
| 5 |
Hb_090530_010 |
0.2324022731 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 6 |
Hb_031658_040 |
0.2381748137 |
- |
- |
- |
| 7 |
Hb_012165_020 |
0.239529821 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |
| 8 |
Hb_001516_050 |
0.2423507 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 9 |
Hb_007313_100 |
0.2425725688 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS2, putative [Ricinus communis] |
| 10 |
Hb_007313_060 |
0.2441365087 |
- |
- |
PREDICTED: paramyosin [Jatropha curcas] |
| 11 |
Hb_000463_030 |
0.2471793953 |
- |
- |
- |
| 12 |
Hb_004785_070 |
0.2479923488 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_024714_080 |
0.2568409136 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 14 |
Hb_008092_080 |
0.2570214279 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 15 |
Hb_006618_050 |
0.2571263611 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_023675_030 |
0.2583731518 |
- |
- |
- |
| 17 |
Hb_003058_020 |
0.2586835711 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_001461_020 |
0.2615345882 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_085083_020 |
0.2648395936 |
- |
- |
PREDICTED: receptor-like protein kinase THESEUS 1 [Jatropha curcas] |
| 20 |
Hb_002495_060 |
0.2654197353 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |