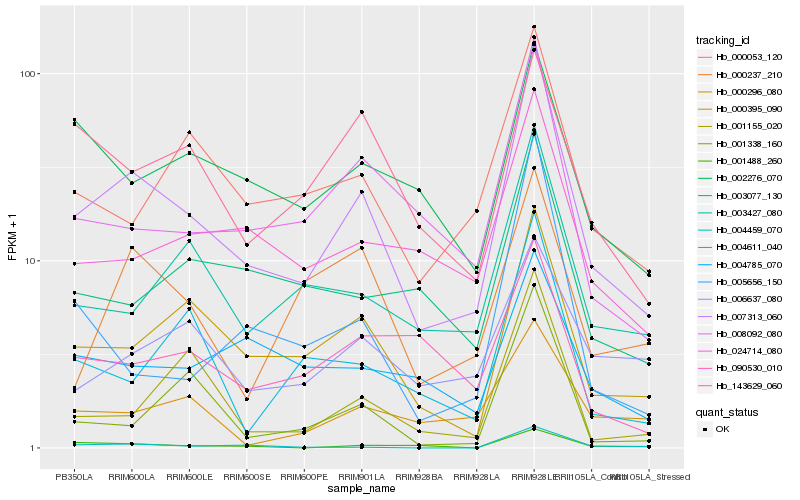
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007313_060 |
0.0 |
- |
- |
PREDICTED: paramyosin [Jatropha curcas] |
| 2 |
Hb_000395_090 |
0.1619174802 |
- |
- |
PREDICTED: uncharacterized protein LOC105645572 [Jatropha curcas] |
| 3 |
Hb_008092_080 |
0.1769954251 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 4 |
Hb_003427_080 |
0.1837109348 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000053_120 |
0.1873321082 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_024714_080 |
0.1917929546 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003077_130 |
0.2076921388 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |
| 8 |
Hb_000296_080 |
0.210040642 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g35130 [Jatropha curcas] |
| 9 |
Hb_004459_070 |
0.2109707455 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 10 |
Hb_004611_040 |
0.2134754808 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigD, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_001488_260 |
0.2157783431 |
- |
- |
Alpha-2-macroglobulin [Gossypium arboreum] |
| 12 |
Hb_006637_080 |
0.217354826 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004785_070 |
0.2186637621 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000237_210 |
0.2189863536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002276_070 |
0.2207424506 |
- |
- |
PREDICTED: heat shock protein 83 [Jatropha curcas] |
| 16 |
Hb_001155_020 |
0.2209480758 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 2 [Theobroma cacao] |
| 17 |
Hb_090530_010 |
0.2228570197 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 18 |
Hb_005656_150 |
0.2231577036 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 19 |
Hb_143629_060 |
0.2235014308 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001338_160 |
0.2246349311 |
- |
- |
NADH dehydrogenase subunit 5 [Hevea brasiliensis] |