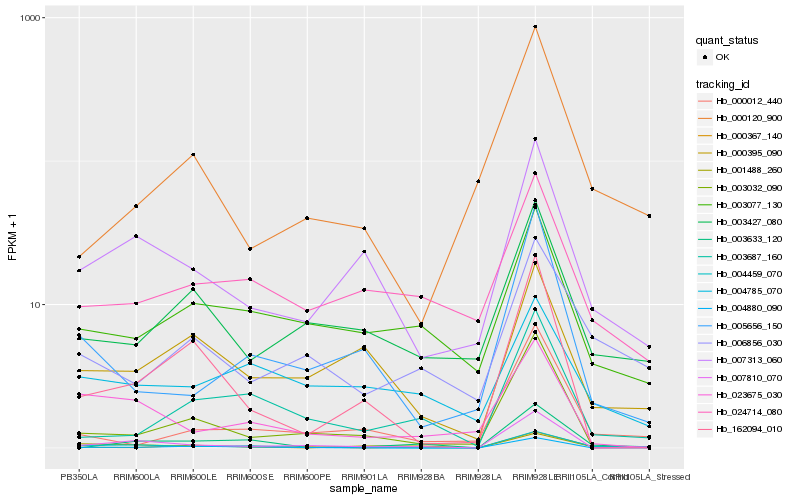
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004459_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 2 |
Hb_007313_060 |
0.2109707455 |
- |
- |
PREDICTED: paramyosin [Jatropha curcas] |
| 3 |
Hb_007810_070 |
0.2364942362 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 4 |
Hb_001488_260 |
0.2400430518 |
- |
- |
Alpha-2-macroglobulin [Gossypium arboreum] |
| 5 |
Hb_005656_150 |
0.2461885647 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 6 |
Hb_023675_030 |
0.2650424217 |
- |
- |
- |
| 7 |
Hb_000395_090 |
0.268154584 |
- |
- |
PREDICTED: uncharacterized protein LOC105645572 [Jatropha curcas] |
| 8 |
Hb_003633_120 |
0.2708044431 |
- |
- |
PREDICTED: uncharacterized protein LOC104880778 [Vitis vinifera] |
| 9 |
Hb_000012_440 |
0.2722705391 |
- |
- |
PREDICTED: uncharacterized protein LOC104248335 [Nicotiana sylvestris] |
| 10 |
Hb_000120_900 |
0.2741480502 |
- |
- |
catalase [Hevea brasiliensis] |
| 11 |
Hb_006856_030 |
0.2751242839 |
- |
- |
Uridine kinase-like protein 4 [Glycine soja] |
| 12 |
Hb_003032_090 |
0.2783797068 |
- |
- |
hypothetical protein POPTR_0018s06450g [Populus trichocarpa] |
| 13 |
Hb_004880_090 |
0.2834772285 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 14 |
Hb_003427_080 |
0.2862044368 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_162094_010 |
0.2877361282 |
- |
- |
RNA polymerase alpha subunit [Hevea brasiliensis] |
| 16 |
Hb_003077_130 |
0.2892149352 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |
| 17 |
Hb_004785_070 |
0.2895285821 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_024714_080 |
0.290223173 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000367_140 |
0.2913994121 |
- |
- |
S-locus-specific glycoprotein S6 precursor, putative [Ricinus communis] |
| 20 |
Hb_003687_160 |
0.2935370031 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |