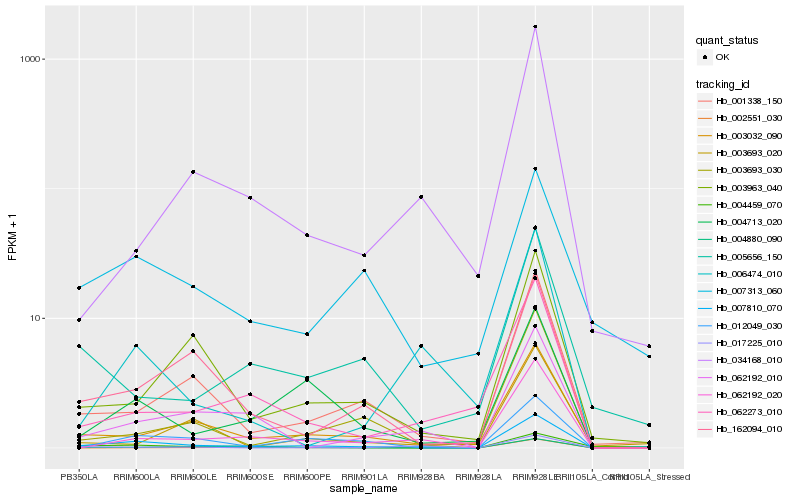
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007810_070 |
0.0 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 2 |
Hb_003032_090 |
0.1859346743 |
- |
- |
hypothetical protein POPTR_0018s06450g [Populus trichocarpa] |
| 3 |
Hb_001338_150 |
0.1982607956 |
- |
- |
hypothetical chloroplast RF1 [Dillenia indica] |
| 4 |
Hb_062192_020 |
0.2129372026 |
- |
- |
cytochrome c biogenesis FN (mitochondrion) [Hevea brasiliensis] |
| 5 |
Hb_004713_020 |
0.2256472489 |
- |
- |
- |
| 6 |
Hb_062192_010 |
0.2281884517 |
- |
- |
- |
| 7 |
Hb_162094_010 |
0.2308945513 |
- |
- |
RNA polymerase alpha subunit [Hevea brasiliensis] |
| 8 |
Hb_003963_040 |
0.2319086978 |
- |
- |
ATP-dependent Clp protease proteolytic subunit [Hevea brasiliensis] |
| 9 |
Hb_062273_010 |
0.2363542201 |
- |
- |
ATP synthase F0 subunit 9 [Chara vulgaris] |
| 10 |
Hb_012049_030 |
0.2364567041 |
- |
- |
RNA polymerase beta subunit [Hevea brasiliensis] |
| 11 |
Hb_004459_070 |
0.2364942362 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 12 |
Hb_007313_060 |
0.2371839792 |
- |
- |
PREDICTED: paramyosin [Jatropha curcas] |
| 13 |
Hb_003693_030 |
0.239783774 |
- |
- |
NADH dehydrogenase subunit 7 [Hevea brasiliensis] |
| 14 |
Hb_002551_030 |
0.2421072321 |
- |
- |
gypsy/Ty-3 retroelement polyprotein; 69905-74404 [Arabidopsis thaliana] |
| 15 |
Hb_034168_010 |
0.2424658841 |
- |
- |
hypothetical protein [Azospirillum sp. CAG:260] |
| 16 |
Hb_004880_090 |
0.243749321 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 17 |
Hb_005656_150 |
0.2472009552 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 18 |
Hb_003693_020 |
0.2479498452 |
- |
- |
Ycf1, partial (chloroplast) [Clusia rosea] |
| 19 |
Hb_006474_010 |
0.248815048 |
- |
- |
hypothetical protein VITISV_032357 [Vitis vinifera] |
| 20 |
Hb_017225_010 |
0.2490752125 |
- |
- |
PREDICTED: nodulation receptor kinase-like [Jatropha curcas] |